|
CGAL 6.1 - Homological Discrete Vector Fields
|
|
CGAL 6.1 - Homological Discrete Vector Fields
|
Classes | |
| class | Abstract_simplicial_chain_complex |
The class Abstract_simplicial_chain_complex represents (topological) chain complexes associated to abstract simplicial complexes. More... | |
| class | Cub_object_io |
The class Cub_object_io is an intermediate IO class, used to load binary volumes and produce cubical complexes. More... | |
| class | Cubical_chain_complex |
The class Cubical_chain_complex represents (topological) chain complexes associated to cubical complexes. More... | |
| class | Duality_cubical_complex_tools |
The class Duality_cubical_complex_tools is dedicated to Alexander duality for 3D binary volumes. More... | |
| class | Duality_simplicial_complex_tools |
The class Duality_simplicial_complex_tools is dedicated to Alexander duality for 3D surface meshes. More... | |
| class | Filtration_core |
The class Filtration_core implements data structures and methods required by the Filtration concept. More... | |
| class | Filtration_lower_star |
The class Filtration_lower_star implements the lower star filtration on a given complex implementing the concept AbstractChainComplex. More... | |
| class | Hdvf |
The class Hdvf implements homology and cohomology computation via homological discrete vector fields (HDVF for short). More... | |
| class | Hdvf_core |
The class Hdvf_core is the core implementation of homological discrete vector fields (HDVF for short). More... | |
| class | Hdvf_duality |
The class Hdvf_duality is the implementation of homological discrete vector fields (HDVF for short) for Alexander duality computation. More... | |
| class | Hdvf_persistence |
The class Hdvf_persistence computes persistent homology using HDVFs. More... | |
| class | Icosphere_object_io |
| struct | IONodeType |
The class IONodeType implements a simple data type used to import vertices coordinates (nodes) in various dimensions. More... | |
| class | Mesh_object_io |
The class Mesh_object_io is an intermediate IO class, used to load triangular/tetraedral meshes and produce simplicial complexes. More... | |
| struct | PairCell |
| Structure to represent data for HDVF operations (pairs of cells). More... | |
| class | Simplex |
The class Simplex is used by the class Abstract_simplicial_chain_complex to implement the structure de simplex (i.e. cells of a simplicial complex). More... | |
| class | Simplicial_chain_complex |
The class Simplicial_chain_complex refines the Abstract_simplicial_chain_complex class by assigning coordinates to vertices (i.e. 0-simplices). More... | |
| class | Sub_chain_complex_mask |
The class Sub_chain_complex_mask is a technical class implementing a sub chain complex. More... | |
| class | Tet_object_io |
| class | Z2 |
The class Z2 implements the concept Ring with the field \(\mathbb Z/2\mathbb Z\). More... | |
| class | Zp |
The class Zp implements the concept Ring with the field \(\mathbb Z/p\mathbb Z\). More... | |
Typedefs | |
| typedef std::vector< size_t > | IOCubCellType |
| Type of cells coordinates in Cub_object_io (Khalimsky or voxel coordinates) | |
| typedef std::vector< IOCubCellType > | IOCubChainType |
| Type of pre-chains in Cub_object_io (list of cells without coefficients). | |
| typedef std::pair< size_t, size_t > | FiltrIndexPerInterval |
| Type to store persistent intervals filtration indices (birth/death indices) | |
| typedef std::pair< size_t, int > | CellDim |
| Type for indexing uniquely a cell. | |
| typedef std::pair< CellDim, CellDim > | CellsPerInterval |
| Type for describing the pair of cells associated to a persistence interval: | |
| template<typename DegType > | |
| using | DegreePerIntervalT = std::pair< DegType, DegType > |
| Template for persistent intervals degrees (birth/death degrees) | |
| template<typename DegType > | |
| using | PerHoleT = std::tuple< FiltrIndexPerInterval, CellsPerInterval, DegreePerIntervalT< DegType > > |
| Template for (full) persistent interval data: | |
| typedef std::set< size_t > | IOCellType |
| Type of cells of Mesh_object_io. | |
| typedef std::vector< IOCellType > | IOChainType |
| Type of pre-chains in Mesh_object_io (list of cells without coefficients). | |
Enumerations | |
| enum | FlagType { PRIMARY , SECONDARY , CRITICAL , NONE } |
| HDVF Enum for the label of cells. More... | |
Functions | |
| ostream & | operator<< (ostream &out, std::vector< size_t > c) |
| template<typename ComplexType > | |
| std::function< double(size_t)> | deg_fun (const ComplexType &complex, std::function< double(const vector< double > &)> &f) |
| Degree function from a coordinates to scalar map. | |
| template<typename CoefType , typename ComplexType , template< typename, int > typename _ChainType = OSM::Sparse_chain, template< typename, int > typename _SparseMatrixType = OSM::Sparse_matrix, typename VertexIdType = size_t> | |
| void | hdvf_geometric_chain_complex_output_vtk (Hdvf_core< CoefType, ComplexType, _ChainType, _SparseMatrixType > &hdvf, ComplexType &complex, string filename="test", bool co_faces=false) |
Exports all the Hdvf information of a geometric chain complex to vtk files. | |
| template<typename CoefType , typename ComplexType , typename DegType , typename FiltrationType > | |
| void | hdvf_persistence_geometric_chain_complex_output_vtk (Hdvf_persistence< CoefType, ComplexType, DegType, FiltrationType > &per_hdvf, ComplexType &complex, string filename="per", bool co_faces=false) |
Exports all the HDVF_persistence information of a geometric chain complex to vtk files. | |
| template<typename CoefType , typename ComplexType , typename VertexIdType = size_t> | |
| void | hdvf_duality_geometric_chain_complex_output_vtk (Hdvf_duality< CoefType, ComplexType > &hdvf, ComplexType &complex, string filename="test", bool co_faces=false) |
Exports all the HDVF_duality information of a geometric chain complex to vtk files. | |
| std::ostream & | operator<< (std::ostream &out, const std::vector< PairCell > &pairs) |
| Overload of operator<< for PairCell type. | |
| template<typename DegType > | |
| ostream & | operator<< (ostream &out, const PerHoleT< DegType > &hole) |
Overload of the << operator to display persistent intervals (that is PerHoleT). | |
| std::ostream & | operator<< (std::ostream &out, const PairCell &p) |
| template<typename CoefType , typename ComplexType > | |
| void | interaction_loop (Hdvf< CoefType, ComplexType > &hdvf, ComplexType &complex, const std::function< void(Hdvf< CoefType, ComplexType > &hdvf, ComplexType &complex)> &output_vtk) |
| Runs an interaction loop to iterated M, W or MW operations and export the results to vtk. | |
| bool | check_sanity_line (const std::string &line, const std::string &file) |
| bool | get_next_uncommented_line (std::ifstream &infile, std::string &result) |
| Mesh_object_io | mesh_BB (const IONodeType &BBmin, const IONodeType &BBmax) |
| size_t | read_nodes (const std::string &node_file, bool load_nodes, std::vector< IONodeType > *nodes) |
Variables | |
| std::function< double(const std::vector< double > &)> | f_x |
| For lower star filtration along x: function mapping coordinates to x. | |
| std::function< double(const std::vector< double > &)> | f_y |
| For lower star filtration along y: function mapping coordinates to y. | |
| std::function< double(const std::vector< double > &)> | f_z |
| For lower star filtration along z: function mapping coordinates to z. | |
| const int | OPT_BND = 0b0001 |
| HDVF option (compute only reduced boundary). | |
| const int | OPT_F = 0b0010 |
| HDVF option (compute only reduced boundary and f). | |
| const int | OPT_G = 0b0100 |
| HDVF option (compute only reduced boundary and g). | |
| const int | OPT_FULL = 0b1000 |
| HDVF option (compute full reduction). | |
| static std::vector< size_t > | VTK_types_IO = {1, 3, 5, 10} |
| typedef std::pair<size_t, int> CGAL::HDVF::CellDim |
Type for indexing uniquely a cell.
"Infinite cells" are defined as (-1,q+1) where q is the dimension of the chain complex.
| typedef std::pair<CellDim, CellDim> CGAL::HDVF::CellsPerInterval |
Type for describing the pair of cells associated to a persistence interval:
For infinite intervals, the "infinite cells" is defined as (-1,q+1) where q is the dimension of the chain complex.
| using CGAL::HDVF::DegreePerIntervalT = typedef std::pair<DegType,DegType> |
Template for persistent intervals degrees (birth/death degrees)
For "infinite" intervals borne at degree d, the interval is set to (d,d-1)
| typedef std::pair<size_t, size_t> CGAL::HDVF::FiltrIndexPerInterval |
Type to store persistent intervals filtration indices (birth/death indices)
For "infinite" intervals borne at index i, the interval is set to (i,i-1)
| using CGAL::HDVF::PerHoleT = typedef std::tuple<FiltrIndexPerInterval, CellsPerInterval, DegreePerIntervalT<DegType> > |
Template for (full) persistent interval data:
| enum CGAL::HDVF::FlagType |
HDVF Enum for the label of cells.
| Enumerator | |
|---|---|
| PRIMARY | |
| SECONDARY | |
| CRITICAL | |
| NONE | |
| void CGAL::HDVF::hdvf_duality_geometric_chain_complex_output_vtk | ( | Hdvf_duality< CoefType, ComplexType > & | hdvf, |
| ComplexType & | complex, | ||
| string | filename = "test", |
||
| bool | co_faces = false |
||
| ) |
Exports all the HDVF_duality information of a geometric chain complex to vtk files.
Export PSC labels and homology/cohomology generators (depending on HDVF options) associated to each persistent intervals to vtk files.
| [in] | hdvf | Reference to the HDVF exported. |
| [in] | complex | Underlying geometric chain complex. |
| [in] | filename | Prefix of all generated files. |
| [in] | co_faces | Export the cohomology generator or its co-faces (sometimes more convenient for visualisation). |
| void CGAL::HDVF::hdvf_geometric_chain_complex_output_vtk | ( | Hdvf_core< CoefType, ComplexType, _ChainType, _SparseMatrixType > & | hdvf, |
| ComplexType & | complex, | ||
| string | filename = "test", |
||
| bool | co_faces = false |
||
| ) |
Exports all the Hdvf information of a geometric chain complex to vtk files.
Export PSC labels and homology/cohomology generators (depending on HDVF options) associated to each critical cell to vtk files.
| [in] | hdvf | Reference to the HDVF exported. |
| [in] | complex | Underlying geometric chain complex. |
| [in] | filename | Prefix of all generated files. |
| [in] | co_faces | Export the cohomology generator or its co-faces (sometimes more convenient for visualisation). |
Below, a sample mesh with, (left) homology generators, (right) two examples of cohomology generators (corresponding generators/co-generators bear similar colours):
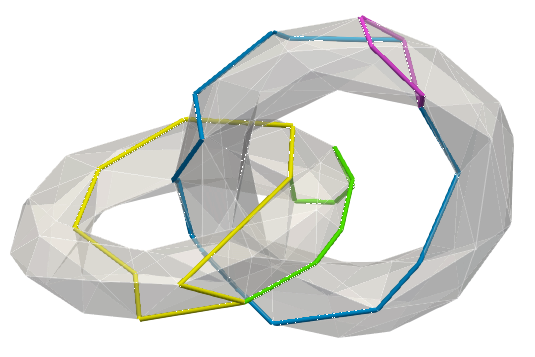
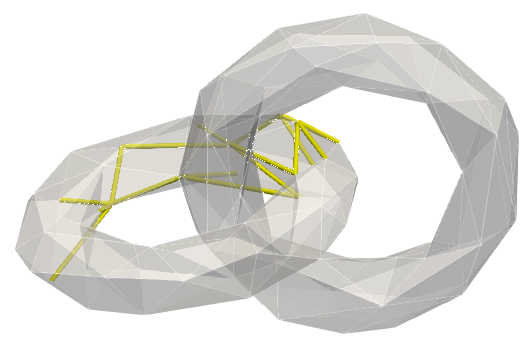

The same generators displayed through their co-faces:


All homology / cohomology generators:
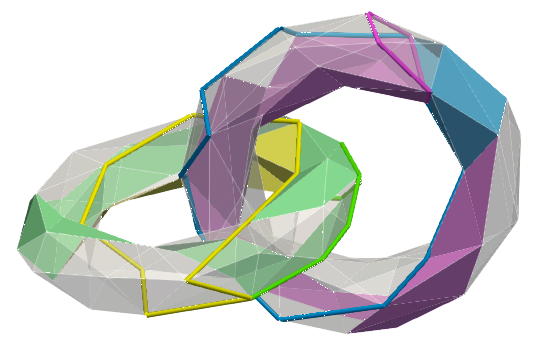
| void CGAL::HDVF::hdvf_persistence_geometric_chain_complex_output_vtk | ( | Hdvf_persistence< CoefType, ComplexType, DegType, FiltrationType > & | per_hdvf, |
| ComplexType & | complex, | ||
| string | filename = "per", |
||
| bool | co_faces = false |
||
| ) |
Exports all the HDVF_persistence information of a geometric chain complex to vtk files.
Export PSC labels and homology/cohomology generators (depending on HDVF options) associated to each persistent intervals to vtk files.
| [in] | per_hdvf | Reference to the persistent HDVF exported. |
| [in] | complex | Underlying geometric chain complex. |
| [in] | filename | Prefix of all generated files. |
| [in] | co_faces | Export the cohomology generator or its co-faces (sometimes more convenient for visualisation). |
| void CGAL::HDVF::interaction_loop | ( | Hdvf< CoefType, ComplexType > & | hdvf, |
| ComplexType & | complex, | ||
| const std::function< void(Hdvf< CoefType, ComplexType > &hdvf, ComplexType &complex)> & | output_vtk | ||
| ) |
Runs an interaction loop to iterated M, W or MW operations and export the results to vtk.
The loop runs until the key Q is pressed. Otherwise, the loop asks for an operation (M, W or MW) and a cell (index and dimension). Then all possible paired cells are listed and the user can chose one of them (or none).
| ostream & CGAL::HDVF::operator<< | ( | ostream & | out, |
| const PerHoleT< DegType > & | hole | ||
| ) |
Overload of the << operator to display persistent intervals (that is PerHoleT).
Format: birth time (cell, dim) -> death time (cell, dim) / degree duration
| std::function<double(const std::vector<double>&)> CGAL::HDVF::f_x |
For lower star filtration along x: function mapping coordinates to x.
| std::function<double(const std::vector<double>&)> CGAL::HDVF::f_y |
For lower star filtration along y: function mapping coordinates to y.
| std::function<double(const std::vector<double>&)> CGAL::HDVF::f_z |
For lower star filtration along z: function mapping coordinates to z.