|
CGAL 6.1 - Homological Discrete Vector Fields
|
|
CGAL 6.1 - Homological Discrete Vector Fields
|
#include <CGAL/HDVF/Hdvf_persistence.h>
CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
The class Hdvf_persistence computes persistent homology using HDVFs.
Hence, unlike other persistence algorithms, beside standard persistent intervals informations (birth/death indices, degrees, associated cells), Hdvf_persistence also provides homology and cohomology generators for persistent pairs. Intuitively, holes die when they are "filled" by a cell: associated homology and cohomology generators provide a representation of the hole and of the cells filling the hole.
Given a Filtration, the Hdvf_persistence constructor basically builds a HDVF where indices of cells in the bases follow the filtration order (permutations between initial indices of cells in the chain complex and new indices given by the filtration are stored). Besides, Hdvf_persistence derives from Hdvf_core with the SparseMatrix parameter type set to Sub_sparse_matrix. Hence, Hdvf_persistence computes homology over a larger and larger sub-complex of K (encoded through a Bitboard mask of Sub_sparse_matrix) following the filtration f. At each step of the filtration, the new cell is paired (A operation) with the youngest cell valid for A.
A few words about persistent generators. At a given time \(t_2\) along the filtration, let us denote by \(\tau\) the \(q+1\)-cell inserted and let us assume that persistent homology computation pairs \(A(\sigma, \tau)\) (let \(t_1\) be the index of \(\sigma\) along the filtration). A new persistent interval \((t_1, t_2)\)is created, generated by \(\sigma\) and filled by \(\tau\). Reduction at time \(t_2-1\) (just before the insertion of \(\tau\)) provides persistent generators: \(g(\sigma)\) is a cycle and \(g(\tau)\) is a chain "filling" this cycle (before the pairing \(A(\sigma, \tau)\), the HDVF over \(K_{t_2}\) is not perfect, thus \(g(\tau)\) is not a cycle but a chain bounded by \(q\)-cycles).
Next figures illustrate such informations for a filtration of a dented double torus.
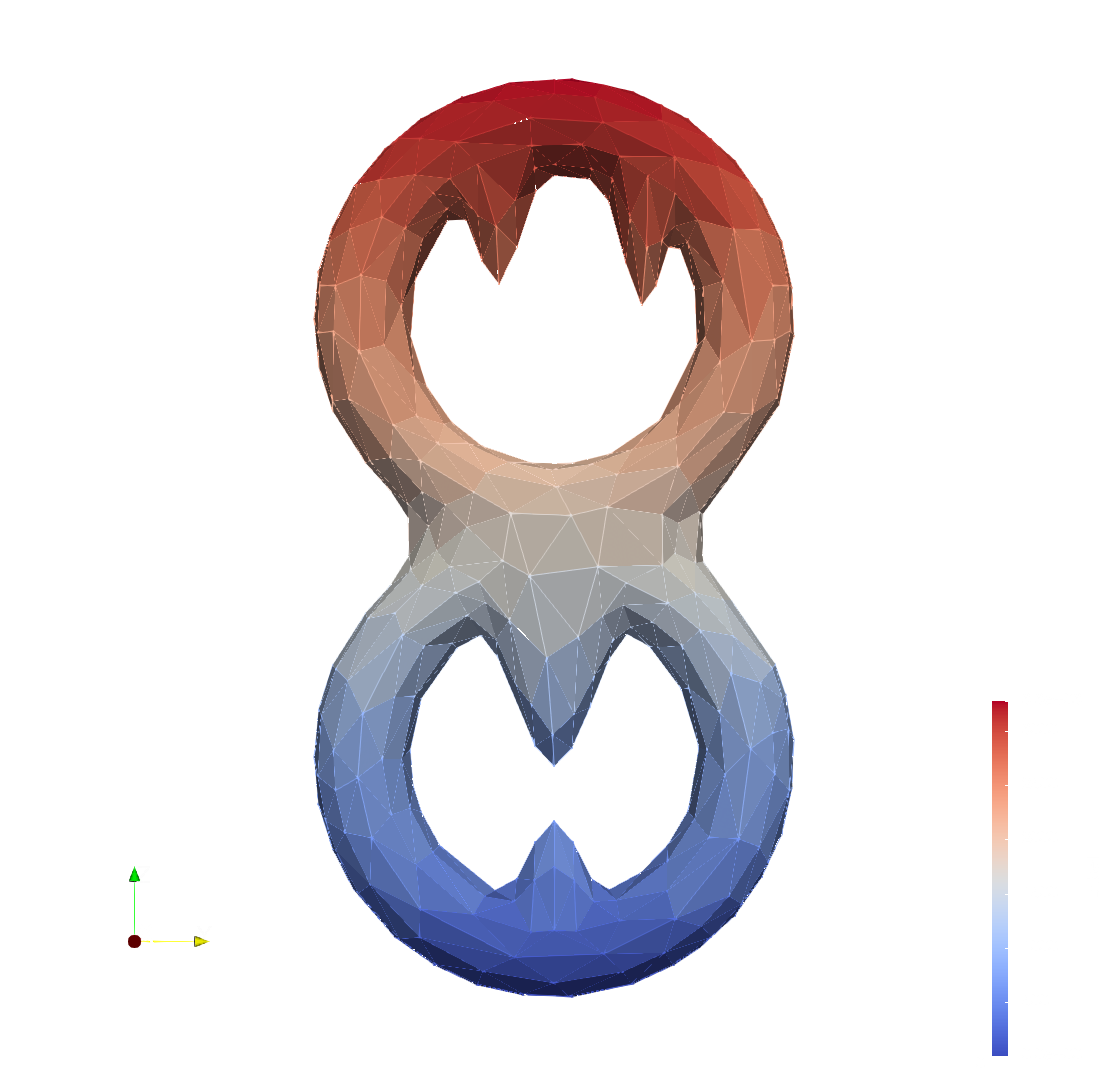
Filtration Filtration_lower_star along the z axis of a dented double torus.
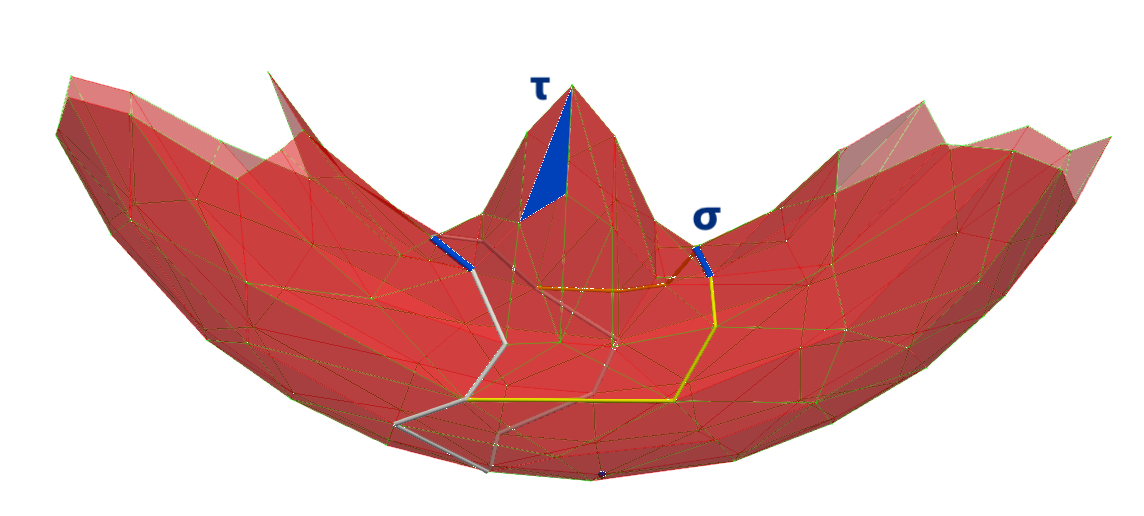

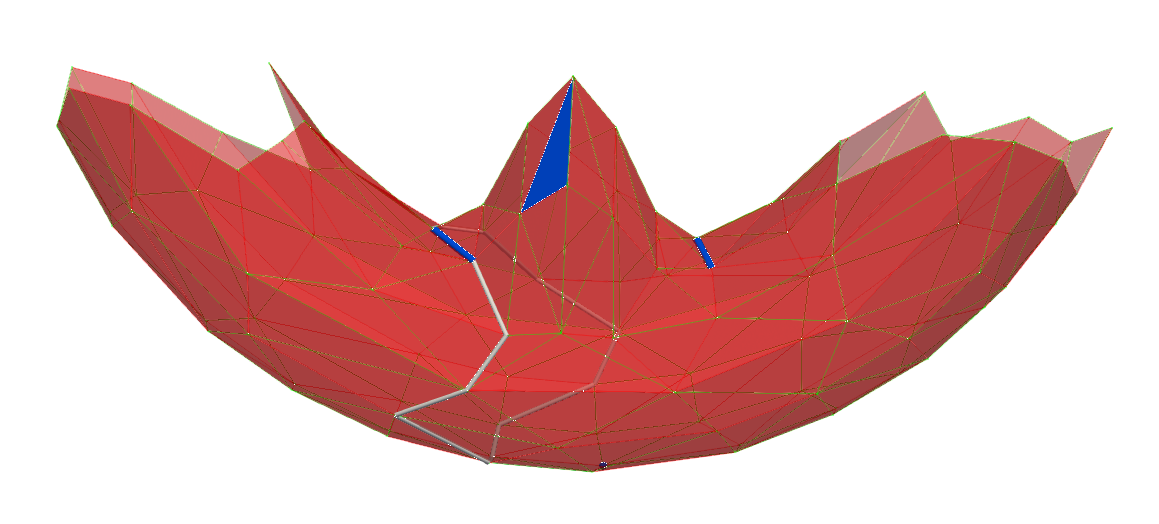
The first persistent hole is a dimension 1 hole with a duration of 0.359 along z.
A(319, 623) between \(\sigma\) and \(\tau\). Middle figure illustrates that in the (non perfect) HDVF before A operation, \(g(\tau)\) is not a cycle but a chain bounded by \(q\)-cycles.A(319, 623) between \(\sigma\) and \(\tau\). Only the white 1-cycle remains (actually its persistent lifetime is infinite).
The figure above illustrates next persistent hole (dimension 0 hole) which duration is 0.684 along z. The cell \(\tau\) is added at time 1237 and fills the 0-hole (connected component) introduced at time 756 by the cell \(\sigma\). The persistent generator associated to \(\sigma\) is simply the cell itself identifying its connected component. The persistent generator associated to \(\tau\) is a 1-chain (depicted in yellow) connecting \(\sigma\) to the critical cell identifying the second connected component.



Figures above illustrate next finite persistent intervals and (right) infinite dimension 1 persistent intervals.
HDVF | CoefficientType | a model of the Ring concept providing the ring used to compute homology. |
| ComplexType | a model of the AbstractChainComplex concept, providing the type of abstract chain complex used. |
| DegType | a scalar data type used for the degrees of the filtration. |
| FiltrationType | a model of the Filtration concept, providing the filtration used to compute persistence. |
Classes | |
| struct | iterator |
| Iterator over (finite) persistent intervals. More... | |
| struct | PerIntervalInformation |
| For persistent diagram iterator (returned by *it) More... | |
Public Types | |
| typedef OSM::Sparse_chain< CoefficientType, OSM::COLUMN > | CChain |
| Type of column-major chains. | |
| typedef OSM::Sparse_chain< CoefficientType, OSM::ROW > | RChain |
| Type of row-major chains. | |
| typedef OSM::Sub_sparse_matrix< CoefficientType, OSM::COLUMN > | CMatrix |
| Type of column-major sparse matrices. | |
| typedef OSM::Sub_sparse_matrix< CoefficientType, OSM::ROW > | RMatrix |
| Type of row-major sparse matrices. | |
| typedef Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | HDVFParent |
| Type of parent HDVF class (Hdvf_core with appropriate template parameters) The SparseMatrix model is set to Sub_sparse_matrix to activate (co)homology computation over a subcomplex. | |
| typedef FiltrationType | Filtration |
| Type of filtrations used to compute persistence. | |
| typedef DegreePerIntervalT< DegType > | DegreePerInterval |
Instanciation of DegreePerIntervalT (persistant intervals degrees) for DegType. | |
| typedef PerHoleT< DegType > | PerHole |
Instanciation of PerHoleT (full persistant intervals informations) for DegType. | |
| typedef std::vector< std::vector< int > > | ExpLabels |
Type for PRIMARY/SECONDARY/CRITICAL labels export Encoding used: | |
| typedef CChain | ExpChain |
| Type chains export (column-major chains) | |
 Public Types inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Public Types inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| typedef OSM::Sparse_chain< CoefficientType, OSM::COLUMN > | CChain |
| Type of column-major chains. | |
| typedef OSM::Sparse_chain< CoefficientType, OSM::ROW > | RChain |
| Type of row-major chains. | |
| typedef OSM::Sub_sparse_matrix< CoefficientType, OSM::COLUMN > | CMatrix |
| Type of column-major sparse matrices. | |
| typedef OSM::Sub_sparse_matrix< CoefficientType, OSM::ROW > | RMatrix |
| Type of row-major sparse matrices. | |
Public Member Functions | |
| Hdvf_persistence (const ComplexType &K, const Filtration &f, int hdvf_opt=OPT_BND, bool with_export=false) | |
| Hdvf_persistence default constructor. | |
| std::vector< PairCell > | compute_perfect_hdvf (bool verbose=false) |
| Computes a perfect persistent HDVF. | |
| bool | with_export () |
| Gets the "with_export" Boolean flag. | |
| const Filtration & | get_filtration () |
| Get a constant reference on the filtration. | |
| DegType | hole_duration (const PerHole hole) const |
| Computes the (degree) duration of a persistent interval (ie. | |
| ostream & | print_hdvf_persistence_info (ostream &out) |
| Prints informations related to the filtration. | |
| virtual vector< vector< int > > | get_psc_labels () const |
| Exports primary/secondary/critical labels (e.g. | |
| virtual CChain | get_homology_chain (size_t cell, int q) const |
Exports homology generators associated to cell (critical cell) of dimension q (e.g. | |
| virtual CChain | get_cohomology_chain (size_t cell, int q) const |
Exports cohomology generators associated to cell (critical cell) of dimension q (used by vtk export). | |
| iterator | begin (bool discard_small=true) |
| Iterator to the beginning of persistent intervals. | |
| iterator | end () |
| Iterator to the ending of the chains indices. | |
 Public Member Functions inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Public Member Functions inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| Hdvf_core (const ComplexType &K, int hdvf_opt=OPT_FULL) | |
| Default constructor. | |
| Hdvf_core (const Hdvf_core &hdvf) | |
| ~Hdvf_core () | |
| virtual PairCell | find_pair_A (int q, bool &found) const |
| Finds a valid PairCell of dimension q / q+1 for A. | |
| virtual PairCell | find_pair_A (int q, bool &found, size_t gamma) const |
Finds a valid PairCell for A containing gamma (a cell of dimension q) | |
| virtual std::vector< PairCell > | find_pairs_A (int q, bool &found) const |
| Finds all valid PairCell of dimension q / q+1 for A. | |
| virtual std::vector< PairCell > | find_pairs_A (int q, bool &found, size_t gamma) const |
Finds all valid PairCell for A containing gamma (a cell of dimension q) | |
| void | A (size_t gamma1, size_t gamma2, int q) |
| A operation: pairs critical cells. | |
| std::vector< PairCell > | compute_perfect_hdvf (bool verbose=false) |
| Computes a perfect HDVF. | |
| std::vector< PairCell > | compute_rand_perfect_hdvf (bool verbose=false) |
| Computes a random perfect HDVF. | |
| bool | is_perfect_hdvf () |
| Tests if a HDVF is perfect. | |
| virtual std::vector< std::vector< size_t > > | get_flag (FlagType flag) const |
Gets cells with a given flag in any dimension. | |
| virtual std::vector< size_t > | get_flag_dim (FlagType flag, int q) const |
Gets cells with a given flag in dimension q. | |
| FlagType | get_cell_flag (int q, size_t tau) const |
Gets the flag of the cell tau in dimension q. | |
| int | get_hdvf_opts () const |
| Gets HDVF computation option. | |
| const RMatrix & | get_f (int q) const |
| Gets the row-major matrix of \(f\) (from the reduction associated to the HDVF). | |
| const CMatrix & | get_g (int q) const |
| Gets the column-major matrix of \(g\) (from the reduction associated to the HDVF). | |
| const CMatrix & | get_h (int q) const |
| Gets the column-major matrix of \(h\) (from the reduction associated to the HDVF). | |
| const CMatrix & | get_dd (int q) const |
| Gets the column-major matrix of \(\partial'\), reduced boundary operator (from the reduction associated to the HDVF). | |
| std::ostream & | print_matrices (std::ostream &out=std::cout) const |
| Prints the matrices of the reduction. | |
| std::ostream & | print_reduction (std::ostream &out=std::cout) const |
| Prints the homology and cohomology reduction information. | |
| virtual std::vector< std::vector< int > > | get_psc_labels () const |
| Exports primary/secondary/critical labels (in particular for vtk export) | |
| virtual CChain | get_homology_chain (size_t cell, int q) const |
Gets homology generators associated to cell (critical cell) of dimension q (used by vtk export). | |
| virtual CChain | get_cohomology_chain (size_t cell, int dim) const |
Gets cohomology generators associated to cell (critical cell) of dimension q (used by vtk export). | |
| std::ostream & | save_hdvf_reduction (std::ostream &out) |
| Saves a HDVF together with the associated reduction (f, g, h, d matrices) | |
| std::istream & | load_hdvf_reduction (std::istream &out) |
| Loads a HDVF together with the associated reduction (f, g, h, d matrices) | |
Protected Attributes | |
| const Filtration & | _f |
| std::vector< std::vector< size_t > > | _K_to_per |
| std::vector< std::vector< size_t > > | _per_to_K |
| std::vector< PerHole > | _persist |
| bool | _with_export |
| std::vector< ExpLabels > | _export_labels |
| std::vector< std::pair< ExpChain, ExpChain > > | _export_g |
| std::vector< std::pair< ExpChain, ExpChain > > | _export_fstar |
| size_t | _t |
| std::vector< size_t > | _t_dim |
| std::vector< OSM::Bitboard > | _masks |
 Protected Attributes inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Protected Attributes inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| std::vector< std::vector< FlagType > > | _flag |
| std::vector< size_t > | _nb_P |
| std::vector< size_t > | _nb_S |
| std::vector< size_t > | _nb_C |
| std::vector< RMatrix > | _F_row |
| std::vector< CMatrix > | _G_col |
| std::vector< CMatrix > | _H_col |
| std::vector< CMatrix > | _DD_col |
| const ComplexType & | _K |
| int | _hdvf_opt |
Friends | |
| ostream & | operator<< (ostream &out, const Hdvf_persistence &per_hdvf) |
| Overload of operator<< for Hdvf_persistence. | |
Additional Inherited Members | |
 Protected Member Functions inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Protected Member Functions inherited from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| OSM::Sparse_chain< CoefficientType, ChainTypeFlag > | projection (const OSM::Sparse_chain< CoefficientType, ChainTypeFlag > &chain, FlagType flag, int q) const |
| void | progress_bar (size_t i, size_t n) |
| typedef std::vector<std::vector<int> > CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::ExpLabels |
Type for PRIMARY/SECONDARY/CRITICAL labels export Encoding used:
PRIMARY: -1SECONDARY: +1CRITICAL: 0 | CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::Hdvf_persistence | ( | const ComplexType & | K, |
| const Filtration & | f, | ||
| int | hdvf_opt = OPT_BND, |
||
| bool | with_export = false |
||
| ) |
Hdvf_persistence default constructor.
Builds an "empty" HDVF_persistence (with all cells critical) associated to the chain complex K and the filtration f. By default, the HDVF option is set to OPT_FULL (full reduction computed)
| [in] | K | A chain complex (a model of AbstractChainComplex). |
| [in] | f | A filtration (a model of Filtration). |
| [in] | hdvf_opt | Option for HDVF computation (OPT_BND, OPT_F, OPT_G or OPT_FULL) |
| [in] | with_export | Boolean option to activate or not the export of PSC labels and homology/cohomology generators for of persistent intervals of positive duration. This information is used by vtk exporters. |
| iterator CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::begin | ( | bool | discard_small = true | ) |
Iterator to the beginning of persistent intervals.
| [in] | discard_small | If true, the iterator visits only persistent intervals of (strictly) positive degree duration. Otherwise, visit all persistent intervals. |
| std::vector< PairCell > CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::compute_perfect_hdvf | ( | bool | verbose = false | ) |
Computes a perfect persistent HDVF.
This method follows the filtration and considers cells one by one. For each of them, it searches the youngest possible cell valid for A (returned by find_pair_A()), and applies the corresponding A() operation. By definition of persistent homology, the Ring of coefficients must be a field.
| [in] | verbose | If this parameter is true, all intermediate reductions are printed out. |
PairCell paired with A. | iterator CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::end | ( | ) |
Iterator to the ending of the chains indices.
|
virtual |
Exports cohomology generators associated to cell (critical cell) of dimension q (used by vtk export).
The method exports the chain \(f^\star(\sigma)\) for \(\sigma\) the cell of index cell and dimension q.
Reimplemented from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Exports homology generators associated to cell (critical cell) of dimension q (e.g.
forx@ vtk export).
The method exports the chain \(g(\sigma)\) for \(\sigma\) the cell of index cell and dimension q.
Reimplemented from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Exports primary/secondary/critical labels (e.g.
for vtk export).
The method exports the labels of every cells in each dimension. Encoding used:
PRIMARY: -1SECONDARY: +1CRITICAL: 0Reimplemented from CGAL::HDVF::Hdvf_core< CoefficientType, ComplexType, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
| DegType CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::hole_duration | ( | const PerHole | hole | ) | const |
Computes the (degree) duration of a persistent interval (ie.
persistent hole)
By definitions of "default" values, infinite intervals have a duration of -1.
| [in] | hole | Persistent interval considered for duration computation |
| ostream & CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::print_hdvf_persistence_info | ( | ostream & | out | ) |
Prints informations related to the filtration.
Prints out the filtration and associated permutations (_K_to_per and _per_to_K) between indices of cells in each dimension in the basis of K and indices along the filtration.
| [in] | out | Reference to an out stream. |
| bool CGAL::HDVF::Hdvf_persistence< CoefficientType, ComplexType, DegType, FiltrationType >::with_export | ( | ) |
Gets the "with_export" Boolean flag.
If the flag is true, homology/cohomology generators and corresponding PSC labels are exported for each persistent interval of positive duration.
|
friend |
Overload of operator<< for Hdvf_persistence.
Prints out finite and infinite persistence intervals.
| [in] | out | Reference to an out stream. |
| [in] | per_hdvf | Constant reference on the Hdvf_persistence to print. |