|
CGAL 6.1 - Homological Discrete Vector Fields
|
|
CGAL 6.1 - Homological Discrete Vector Fields
|
#include <CGAL/HDVF/Cubical_chain_complex.h>
The class Cubical_chain_complex represents (topological) chain complexes associated to cubical complexes.
A cubical complex is a set of "square" cells such that: all the faces of a given cell also belong to the complex and any two cells intersect exactly along a common face.
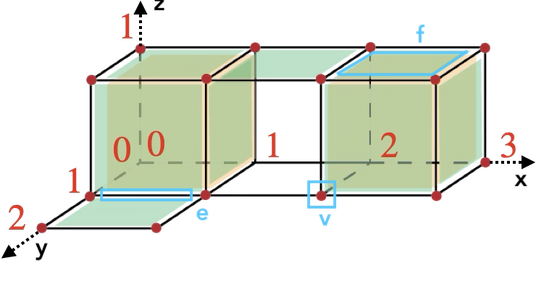
A cell in a cubical complex of dimension q is the cartesian product of \(q\) intervals \([n_1,n_1+\delta_1]\times [n_q,n_q+\delta_q] \) with \(n_i\in \mathbb N \) and \(\delta_i = 0,1\). The dimension of such a cell is \(\sum_{i=1}^q \delta_i\). A 0-cell is thus a vertex, a 1-cell contains one non zero delta (edge along one of the axes), a 2-cell contains two non zero deltas (square along two of the axes), while a 3-cell is a cube...
For instance, \(v = [2,2]\times[1,1]\times[0,0]\), \(e = [0,1]\times[1,1]\times[0,0]\) and \(f = [2,3]\times[0,1]\times[1,1]\).
Given a cubical complex of dimension \(q\) included in \([0,B_1]\times\cdots \times[0,B_q]\), Khalimsky coordinates associate to each cell of the complex a unique coordinate in \([0,2B_1]\times\cdots \times[0,2B_q]\):
\[ \begin{array}{rcl} \sigma & \mapsto & \mathrm{khal}(\sigma) \\ [n_1,n_1+\delta_1]\times\cdots \times [n_q,n_q+\delta_q] & \,\to\, & (2n_1+\delta_1,\ldots,2n_1+\delta_1) \\ \end{array}\]
we set \(N_i = 2B_i+1\) the size of the Khalimsky bounding box along the \(i\)th axis.
The dimension of a cell is the given by the number of odd coordinates. In our previous example, \(B_1=3, B_2=2, B_3=1\), hence \(N_1=7, N_2=5, N_3=3\) and we get the following Khalimsky coordinates: \(\mathrm{khal}(v) = (4,2,0)\), \(\mathrm{khal}(e) = (1,2,0)\) and \(\mathrm{khal}(f) = (5,1,2)\).
The boundary map of the complex is computed by the constructor of the class using the standard formula with Khalimsky coordinates. Given a cell \(\sigma\) with \(\mathrm{khal}(\sigma) = (x_1,\ldots,x_q)\)
\[ \partial(x_1,\ldots,x_q) = \sum_{\substack{i=0\\x_i\text{ odd}}}^q (-1)^{j(i)} \left( (x_1,\ldots,x_i+1,\ldots, x_q) - (x_1,\ldots,x_i-1,\ldots, x_q)\right)\]
where \(j(i)\) is the number of odd coordinates between \(x_1\) and \(x_i\).
Let us also point out that besides Khalimsky coordinates, cells are indexed along each dimension, thus each cell is uniquely determined by its dimension and its index in this dimension (called "base index").
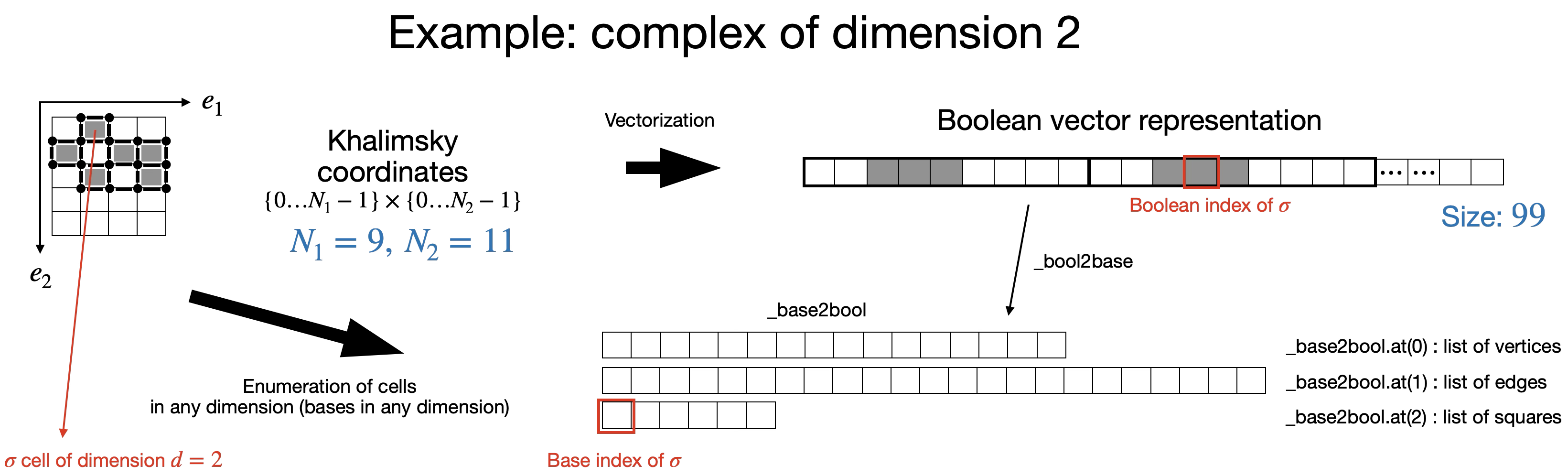
As described above, Khalimsky coordinates provide a convenient tool to identify cells of any dimension. Hence a complex of dimension \(q\) can be encoded by a boolean matrix of size \(N_1\times\cdots\times N_q\) (with previous notations). For convenience, this matrix is vectorized and thus the complex is stored in a boolean vector (denoted by _cells) of size \(N_1\times \cdots \times N_q\).
Cells of any dimension are thus repesented by a given element of this boolean vector and the corresponding index is called their boolean index.
As stated above, besides this boolean representation, topological computations require to identify the bases of cells in any dimension (bases of the free chain groups). Hence, a cell of dimension \(d\) is also identified by its index in the basis of \(d\)-dimensional cells. This index is called its basis index. The vector _bool2base stores, for each dimension, the map between boolean and base indices, while the _base2bool stores, for each dimension, the permutation between base and boolean indices.
GeometricChainComplex | CoefficientType | a model of the Ring concept. |
Public Types | |
| enum | typeComplexCube { PRIMAL , DUAL } |
| Type used to encode primal or dual construction. More... | |
| typedef std::vector< double > | Point |
| Type of vertices coordinates. | |
| typedef OSM::Sparse_chain< CoefficientType, OSM::COLUMN > | CChain |
| Type of column-major chains. | |
| typedef OSM::Sparse_chain< CoefficientType, OSM::ROW > | RChain |
| Type of row-major chains. | |
| typedef OSM::Sparse_matrix< CoefficientType, OSM::COLUMN > | CMatrix |
| Type of column-major sparse matrices. | |
Public Member Functions | |
| Cubical_chain_complex () | |
| Default constructor (empty cubical complex). | |
| Cubical_chain_complex (const Cub_object_io &cub, typeComplexCube type) | |
Constructor from a Cub_object_io (builds PRIMAL or DUAL associated complex depending on type). | |
| Cubical_chain_complex & | operator= (const Cubical_chain_complex &complex) |
| Assignment operator for cubical chain complexes. | |
| CChain | d (size_t id_cell, int q) const |
| Returns the boundary of the cell id_cell in dimension q. | |
| RChain | cod (size_t id_cell, int q) const |
| Returns the co-boundary of the cell id_cell in dimension q. | |
| int | dim () const |
| Returns the dimension of the complex. | |
| size_t | nb_cells (int q) const |
| Returns the number of cells in a given dimension. | |
| const vector< CMatrix > & | get_bnd_matrices () const |
| Returns a constant reference to the vector of boundary matrices (along each dimension). | |
| const CMatrix & | get_bnd_matrix (int q) const |
| Returns a copy of the dim-th boundary matrix (ie. | |
| std::vector< size_t > | bottom_faces (size_t id_cell, int q) const |
| Returns dimension 0 cells indexes included in the cell with index id_cell of dimension q. | |
| template<typename CoefficientT , int ChainTypeF> | |
| CChain | cofaces_chain (OSM::Sparse_chain< CoefficientT, ChainTypeF > chain, int q) const |
Returns the cofaces of a given chain in dimension q. | |
| void | print_complex () const |
| Prints informations on the complex. | |
| size_t | get_id () const |
| Gets (unique) object Id. | |
| Point | get_vertex_coords (size_t i) const |
| Gets the coordinates of the ith vertex. | |
| const std::vector< Point > & | get_vertices_coords () const |
| Gets the vector of vertices coordinates | |
Static Public Member Functions | |
| template<typename LabelType = int> | |
| static void | chain_complex_to_vtk (const Cubical_chain_complex< CoefficientType > &K, const std::string &filename, const std::vector< std::vector< LabelType > > *labels=NULL, string label_type_name="int") |
| END Methods of the Cubical_chain_complex concept. | |
| static void | chain_complex_chain_to_vtk (const Cubical_chain_complex< CoefficientType > &K, const std::string &filename, const OSM::Sparse_chain< CoefficientType, OSM::COLUMN > &chain, int q, size_t cellId=-1) |
| Exports a chain over a cubical complex to a VTK file. | |
Public Attributes | |
| friend | Duality_cubical_complex_tools< CoefficientType > |
Friend class Duality_cubical_complex_tools provides tools for Alexander duality. | |
Protected Member Functions | |
| std::vector< size_t > | get_size_bb () const |
| std::vector< size_t > | get_P () const |
| std::vector< bool > | get_cells () const |
| std::vector< std::vector< size_t > > | get_base2bool () const |
| std::vector< std::map< size_t, size_t > > | get_bool2base () const |
| CChain | boundary_cell (size_t index_base, int dim) const |
| bool | is_valid_cell (const std::vector< size_t > &cell) const |
| bool | is_valid_cell (size_t id_cell) const |
| vector< size_t > | khal_to_verts (vector< size_t > c) const |
| void | initialize_cells (const Cub_object_io &cub, typeComplexCube type) |
| std::vector< size_t > | ind2khal (size_t index) const |
| Computes Khalimsky coordinates from boolean index. | |
| size_t | khal2ind (const std::vector< size_t > &base_indices) const |
| std::vector< size_t > | ind2vox (size_t index, vector< size_t > B, size_t max_size) const |
| size_t | vox2ind (const std::vector< size_t > &base_indices, vector< size_t > B, size_t max_size) const |
| void | calculate_d (int q) |
| void | insert_cell (size_t cell) |
| int | calculate_dimension (const std::vector< size_t > &cell) const |
| int | calculate_dimension (size_t cell_index) const |
| std::vector< size_t > | calculate_boundaries (size_t cell) const |
Protected Attributes | |
| int | _dim |
| std::vector< size_t > | _size_bb |
| std::vector< size_t > | _P |
| std::vector< bool > | _cells |
| std::vector< std::vector< size_t > > | _base2bool |
| std::vector< std::map< size_t, size_t > > | _bool2base |
| std::vector< CMatrix > | _d |
| enum CGAL::HDVF::Cubical_chain_complex::typeComplexCube |
| CGAL::HDVF::Cubical_chain_complex< CoefficientType >::Cubical_chain_complex | ( | ) |
Default constructor (empty cubical complex).
Builds an empty cubical complex.
| CGAL::HDVF::Cubical_chain_complex< CoefficientType >::Cubical_chain_complex | ( | const Cub_object_io & | cub, |
| typeComplexCube | type | ||
| ) |
Constructor from a Cub_object_io (builds PRIMAL or DUAL associated complex depending on type).
Builds the cubical complex associated to a a set of cells (vertices, edges, squares, cubes...), ie. performs the down closure of cells and set the boundary matrices in any dimension. Given a set of cells:
type is PRIMAL, the constructor builds the associated complex as such (see below middle), which comes to encode \(3^q-1\) connectivity (with \(q\) the dimension of the complex)type is DUAL and the Cub_object_io contains only cells of maximal dimension (ie. binary object), the constructor build the dual associated complex (see below right), which comes to encode \(2q\) connectivity (with \(q\) the dimension of the complex)
| [in] | cub | A Cub_object_io containing a set of "cubical" cells. |
| [in] | type | Type of construction used (PRIMAL or DUAL). |
| std::vector< size_t > CGAL::HDVF::Cubical_chain_complex< CoefficientType >::bottom_faces | ( | size_t | id_cell, |
| int | q | ||
| ) | const |
Returns dimension 0 cells indexes included in the cell with index id_cell of dimension q.
Returns the dimension 0 vertices indexes included in the cell with index id_cell of dimension q.
| [in] | id_cell | Index of the cell. |
| [in] | q | Dimension of the cell. |
|
static |
Exports a chain over a cubical complex to a VTK file.
The method generates legacy text VTK files. All the cells of the chain with non zero coefficient are exported. If a cellId is provided, labels are exported in a VTK property (2 for all cells, 0 for cell of index cellId). The index of each cell is exported in a CellID property.
| [in] | K | Cubical complex exported. |
| [in] | filename | Output file root (output filenames will be built from this root). |
| [in] | chain | Sparse_chain exported (all the cells with non-zero coefficients in the chain are exported to vtk). |
| [in] | q | Dimension of the cells of the chain. |
| [in] | cellId | If different from MAX_SIZE_T, labels are exported to distinguish cells of the chain (label 2) from cellId cell (label 0). |
|
static |
END Methods of the Cubical_chain_complex concept.
Exports a cubical complex (plus, optionally, labels) to a VTK file.
The method generates legacy text VTK files. Labels are exported as such in a VTK property, together with CellID property, containing the index of each cell.
| LabelType | Type of labels provided (default: int). |
| [in] | K | Cubical complex exported. |
| [in] | filename | Output file root (output filenames will be built from this root). |
| [in] | labels | Pointer to a vector of labels in each dimension. (*labels).at(q) is the set of integer labels of cells of dimension q. If labels is NULL, only CellID property is exported. |
| [in] | label_type_name | Typename used in vtk export (e.g. "int" or "unsigned_long", see VTK manual ). |
| RChain CGAL::HDVF::Cubical_chain_complex< CoefficientType >::cod | ( | size_t | id_cell, |
| int | q | ||
| ) | const |
Returns the co-boundary of the cell id_cell in dimension q.
Returns a row-major chain containing the co-boundary of the cell id_cell in dimension q (so actually a row of the boundary matrix).
| [in] | id_cell | Index of the cell. |
| [in] | q | Dimension of the cell. |
| CChain CGAL::HDVF::Cubical_chain_complex< CoefficientType >::cofaces_chain | ( | OSM::Sparse_chain< CoefficientT, ChainTypeF > | chain, |
| int | q | ||
| ) | const |
Returns the cofaces of a given chain in dimension q.
The resulting chain lies in dimension q+1 and is null if this dimension exceeds the dimension of the complex.
| CChain CGAL::HDVF::Cubical_chain_complex< CoefficientType >::d | ( | size_t | id_cell, |
| int | q | ||
| ) | const |
Returns the boundary of the cell id_cell in dimension q.
Returns a copy of the column-major chain stored in the boundary matrix of dimension q: boundary of the cell id_cell in dimension q.
| [in] | id_cell | Index of the cell. |
| [in] | q | Dimension of the cell. |
| int CGAL::HDVF::Cubical_chain_complex< CoefficientType >::dim | ( | ) | const |
Returns the dimension of the complex.
Returns the dimension of the cubical complex.
| const vector< CMatrix > & CGAL::HDVF::Cubical_chain_complex< CoefficientType >::get_bnd_matrices | ( | ) | const |
Returns a constant reference to the vector of boundary matrices (along each dimension).
Returns a constant reference to the vector of boundary matrices along each dimension. The q-th element of this vector is a column-major sparse matrix containing the boundaries of q-cells (ie. rows encode q-1 cells and columns q cells).
| const CMatrix & CGAL::HDVF::Cubical_chain_complex< CoefficientType >::get_bnd_matrix | ( | int | q | ) | const |
Returns a copy of the dim-th boundary matrix (ie.
column-major matrix of \(\partial_q\)).
It is a column-major sparse matrix containing the boundaries of dim-cells (ie. rows encode q-1 cells and columns q cells).
| [in] | q | Dimension of the boundary matrix (ie. columns will contain the boundary of dimension q cells). |
| size_t CGAL::HDVF::Cubical_chain_complex< CoefficientType >::get_id | ( | ) | const |
Gets (unique) object Id.
For comparison of constant references to the complex.
| size_t CGAL::HDVF::Cubical_chain_complex< CoefficientType >::nb_cells | ( | int | q | ) | const |
Returns the number of cells in a given dimension.
| [in] | q | Dimension along which the number of cells is returned. |
| Cubical_chain_complex & CGAL::HDVF::Cubical_chain_complex< CoefficientType >::operator= | ( | const Cubical_chain_complex< CoefficientType > & | complex | ) |
Assignment operator for cubical chain complexes.
Stores a copy of an cubical chain complex in *this.
| [in] | complex | The cubical chain complex which will be copied. |
| void CGAL::HDVF::Cubical_chain_complex< CoefficientType >::print_complex | ( | ) | const |
Prints informations on the complex.
Displays the number of cells in each dimension and the boundary matrix in each dimension.