|
CGAL 6.2 - Homological Discrete Vector Fields
|
|
CGAL 6.2 - Homological Discrete Vector Fields
|
The concept HDVF describes the requirements for Homological Discrete Vector Fields (HDVF for short) , a theory of computational homology unifying discrete Morse theory and effective homology. HDVFs were introduced by Aldo Gonzalez-Lorenzo in his PhD (see [AGL,2017], [AGL,2016]).
A HDVF is a combinatorial tool computing the homology of a chain complex (described by the AbstractChainComplex concept) including "geometric" information such as homology and cohomology generators.
As described in the user manual description, an HDVF is:
More precisely, a HDVF over a chain complex C, derived from a complex K, is a partition of the cells: \(K = P\coprod S\coprod C\) with \(P\) primary cells, \(S\) secondary cells and \(C\) critical cells, such that the sub-matrix:
\[\partial (S)|_P\text{ is invertible.} \]
In what follows, given \(q\in \mathbb N\) and \(G\) a sub-group of the qth chain group \(C_q\), we denote by:
That is, for all \(q\in \mathbb N\), if
\[\mathrm{Mat}(\partial_q) = \begin{array}{lc} & P_q \ \ \ \ S_q \ \ \ \ C_q \\ \begin{array}{l} P_{q-1} \\ S_{q-1} \\ C_{q-1} \\ \end{array} & \left(\begin{array}{ccc} \star & D_{PS} & \star \\ \star & \star & \star \\ \star & \star & \star \\ \end{array}\right)\\ \end{array} \]
then \(D_{PS}\), the matrix of \(j_P\circ \partial\circ i_S\), also written \(\partial (S)|_P\) is invertible.
In order to build a perfect HDVF, a pairing operation called A() associates (under conditions) two critical cells (becoming PRIMARY and SECONDARY) and updates the reduction in time \(\mathscr O(n^2)\). Intuitively, this operation bears some similitudes (see users guide) with discrete Morse theory's DGVF arrows (and an HDVF can always be represented by such a vector field - possibly with cycles). Given a HDVF \(X = (P,S,C)\), A operation between two cells \((\gamma_1, \gamma_2)\) of respective dimension \(q/q+1\) is valid if \((P',S',C') = (P\cup\{\gamma_1\}, P\cup\{\gamma_2\}, C\backslash\{\gamma_1,\gamma_2\})\) is a HDVF (ie. if \(j_{P'}\circ \partial\circ i_{S'}\) is still invertible). This is actually the case if and only if \(\langle d(\gamma_2), \gamma_1 \rangle\) (with \(d\) the reduced boundary operator) is invertible. This condition is called the validity condition for A; all the find_pair_A() and find_pairs_A() methods search for such valid pairs.
Starting from this operation, HDVF provides two methods to generate perfect HDVFs (and hence compute homology):
compute_perfect_hdvf() which computes a perfect HDVF by chosing at each step the first A-pairings available (depending on cells ordering in the chain complex)compute_rand_perfect_hdvf() which computes a perfect random HDVF by chosing at each step a rand A-pairing among all possible ones (this option is thus slower)For efficiency, the HDVF reduction can be computed partially:
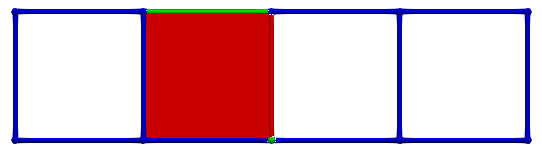
OPT_BND): to get Betti numbersOPT_G): to get homology generatorsOPT_F): to get co-homology generatorsOPT_FULL): to get the full homological / cohomological informationLet us consider a simple cubical complex example (see Cubical chain complexes for an introduction). In what follows, PRIMARY cells are coloured green, SECONDARY are coloured red and critical blue. The picture below shows an initial empty HDVF where all cells are critical:
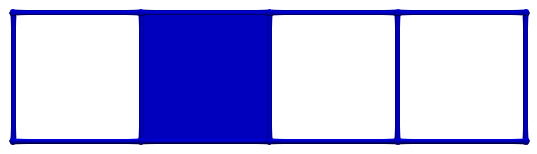
The boundary matrix of \(\partial_2\) (which equals the reduced boundary matrix as the HDVF is empty) for \(\mathbb Z\)-homology is (cells are identified by their Khalimsky coordinates):
\[\mathrm{Mat}(\partial_2) = \begin{array}{lc} & (3,1)\\ \begin{array}{l} (3,0) \\ (2,1) \\ (4,1) \\ (3,2) \\ \end{array} & \left(\begin{array}{c} 1 \\ -1 \\ 1 \\ -1 \\ \end{array}\right)\\ \end{array} \]
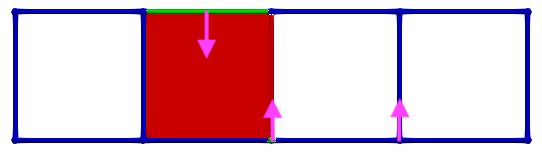
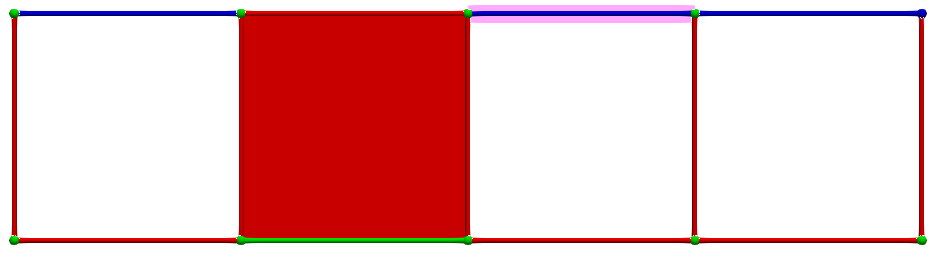
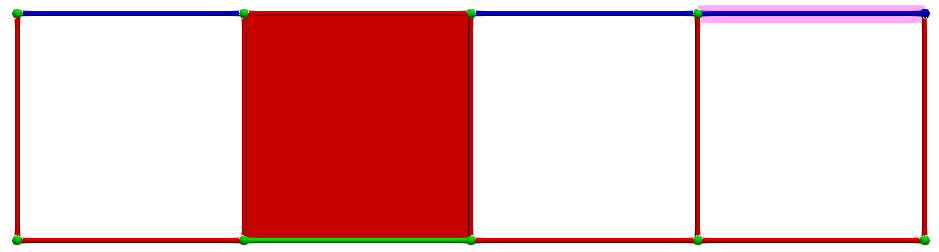
Let us consider \(\gamma_2 = (3,1)\) and \(\gamma_1 = (3,2)\), the condition for A is valid (-1 is invertible). The following images illustrates this pairing in discrete Morse theory "style" (pink arrow in left complex) and the resulting HDVF (right complex).
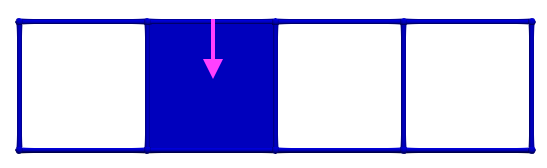
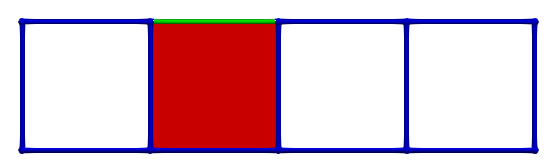
The reduced boundary matrix in dimension 2 becomes empty and the matrix of \(d_1\) is similar to the boundary matrix \(\partial_1\) with the row/colums associated to previous cells removed.
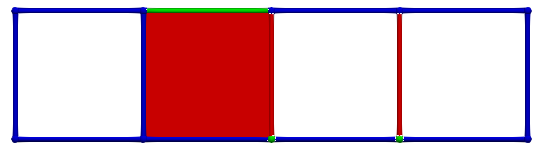
Thus, if we now consider \(\gamma_2 = (4,1)\) and \(\gamma_1 = (4,0)\), the condition for A is valid; cells can be paired and we obtain the following HDVF:
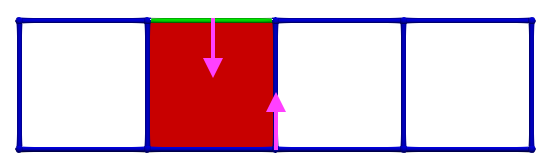
The process can be continued step by step:
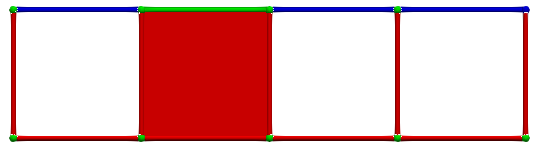
Until we eventually get a perfect HDVF:
Each hole is identified by a critical cell and the \(g\) map of the reduction provides associated homology generators (highlighted in blue):
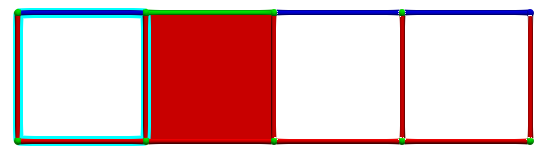
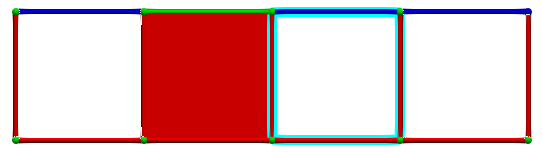
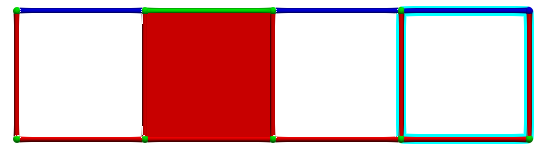
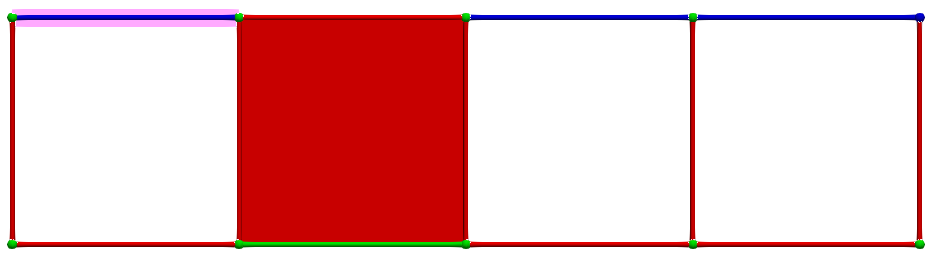
A corresponding cohomology generators (highlighted in pink):
CGAL::Homological_discrete_vector_field::Hdvf_core<IntegralDomainWithoutDivision, AbstractChainComplex, SparseChain, Sparse_matrix> CGAL::Homological_discrete_vector_field::Hdvf<IntegralDomainWithoutDivision, AbstractChainComplex> CGAL::Homological_discrete_vector_field::Hdvf_persistence<IntegralDomainWithoutDivision, AbstractChainComplex> CGAL::Homological_discrete_vector_field::Hdvf_duality<IntegralDomainWithoutDivision, AbstractChainComplex> How to describe constants declared in the namespace Homological_discrete_vector_field and used everywhere? PSC_flag, options, exporttype*
[AGL, 2017] Aldo Gonzalez-Lorenzo, Alexandra Bac, Jean-Luc Mari, Pedro Real. Allowing cycles in discrete Morse theory, Topology and its Applications, Volume 228, 2017, Pages 1-35.
[AGL, 2016] Aldo Gonzalez-Lorenzo. Computational Homology Applied to Discrete Objects. Discrete Mathematics [cs.DM]. Aix-Marseille Université; Universidad de Sevilla, 2016.
Concepts | |
| concept | Cell_pair |
| Structure to represent data for HDVF operations (pairs of cells). More... | |
Types | |
| typedef unspecified_type | Complex |
Type of underlying chain complex (a model of AbstractChainComplex). | |
| typedef Complex::Coefficient_ring | Coefficient_ring |
| Type of coefficients used to compute homology. | |
| typedef unspecified_type | Sparse_chain |
Type of sparse chains (a model of SparseChain). | |
| typedef unspecified_type | Sparse_matrix |
Type of sparse matrices (a model of SparseMatrix). | |
| typedef Sparse_chain< CoefficientType, CGAL::OSM::COLUMN > | Column_chain |
| Type of column-major chains. | |
| typedef Sparse_chain< CoefficientType, CGAL::OSM::ROW > | Row_chain |
| Type of row-major chains. | |
| typedef Sparse_matrix< CoefficientType, CGAL::OSM::COLUMN > | Column_matrix |
| Type of column-major sparse matrices. | |
| typedef Sparse_matrix< CoefficientType, CGAL::OSM::ROW > | Row_matrix |
| Type of row-major sparse matrices. | |
Functions to find valid pairs for <tt>A()</tt> operations | |
| virtual Cell_pair | find_pair_A (int q, bool &found) const |
| Finds a valid Cell_pair of dimension q / q+1 for A. | |
| virtual Cell_pair | find_pair_A (int q, bool &found, size_t gamma) const |
Finds a valid Cell_pair for A containing gamma (a cell of dimension q) | |
| virtual std::vector< Cell_pair > | find_pairs_A (int q, bool &found) const |
| Finds all valid Cell_pair of dimension q / q+1 for A. | |
| virtual std::vector< Cell_pair > | find_pairs_A (int q, bool &found, size_t gamma) const |
Finds all valid Cell_pair for A containing gamma (a cell of dimension q) | |
HDVF functions and operations | |
| void | A (size_t gamma1, size_t gamma2, int q) |
| A operation: pairs two critical cells. | |
| std::vector< Cell_pair > | compute_perfect_hdvf (bool verbose=false) |
| Computes a perfect HDVF. | |
| std::vector< Cell_pair > | compute_rand_perfect_hdvf (bool verbose=false) |
| Computes a random perfect HDVF. | |
Getters | |
| std::vector< std::vector< size_t > > | flag (PSC_flag flag) const |
Gets cells with a given PSC_flag in any dimension. | |
| std::vector< size_t > | psc_flags (PSC_flag flag, int q) const |
Gets cells with a given PSC_flag in dimension q. | |
| PSC_flag | psc_flag (int q, size_t tau) const |
Gets the PSC_flag of the cell tau in dimension q. | |
| int | hdvf_opts () |
| Gets HDVF option. | |
| const Row_matrix & | matrix_f (int q) const |
| Gets the row-major matrix of \(f\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_g (int q) const |
| Gets the column-major matrix of \(g\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_h (int q) const |
| Gets the column-major matrix of \(h\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_dd (int q) const |
| Gets the column-major matrix of \(\partial'\), reduced boundary operator (from the reduction associated to the HDVF). | |
| bool | is_perfect_hdvf () |
| Tests if a HDVF is perfect. | |
Output functions | |
| std::ostream & | write_matrices (std::ostream &out=std::cout) const |
| Writes the matrices of the reduction. | |
| std::ostream & | write_reduction (std::ostream &out=std::cout) const |
| Writes the homology and cohomology reduction information. | |
| std::ostream & | write_hdvf_reduction (std::ostream &out) |
| Writes a HDVF and its reduction to a stream. | |
| std::istream & | read_hdvf_reduction (std::istream &in) |
| Reads a HDVF and its reduction from a stream. | |
| virtual std::vector< std::vector< int > > | psc_labels () const |
| Exports primary/secondary/critical labels (in particular for vtk export). | |
| virtual Column_chain | homology_chain (size_t cell_index, int q) const |
Returns a chain containing the homology generator associated to cell_index (critical cell) of dimension q (in particular for vtk export). | |
| virtual Column_chain | cohomology_chain (size_t cell_index, int dim, bool co_faces=false) const |
Returns a chain containing the cohomology generators associated to cell_index (critical cell) of dimension q (in particular for vtk export). | |
| void HDVF::A | ( | size_t | gamma1, |
| size_t | gamma2, | ||
| int | q | ||
| ) |
A operation: pairs two critical cells.
A pair of critical cells \((\gamma_1, \gamma_2)\) of respective dimension q and q+1 is valid for A if \(\langle \partial_{q+1}(\gamma_2), \gamma_1 \rangle\) is invertible. After the A() operation, \(\gamma_1\) becomes PRIMARY, \(\gamma_2\) becomes SECONDARY. The A functions updates the reduction accordingly (in time \(\mathscr O(n^2)\)).
|
virtual |
Returns a chain containing the cohomology generators associated to cell_index (critical cell) of dimension q (in particular for vtk export).
The method exports:
cell_index and dimension q,co_faces is true (sometimes easier to view cohomology generators)Below, a sample mesh with, (left) homology generators, (right) two examples of cohomology generators (corresponding generators/co-generators bear similar colours):
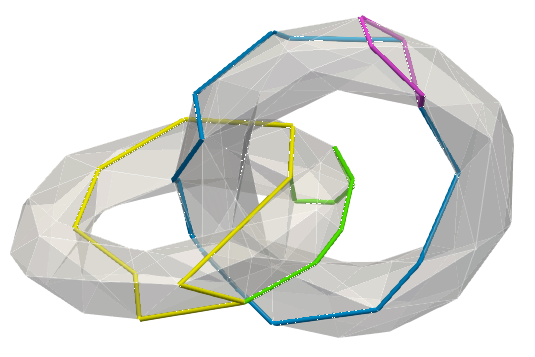
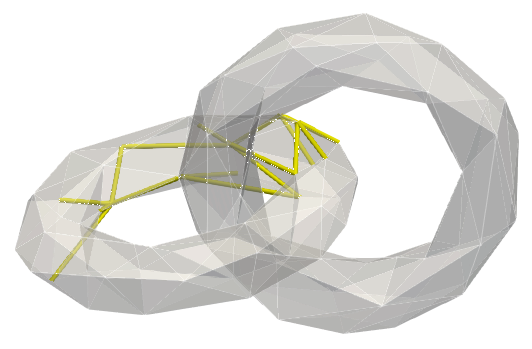

The same generators displayed through their co-faces:


All homology / cohomology generators:
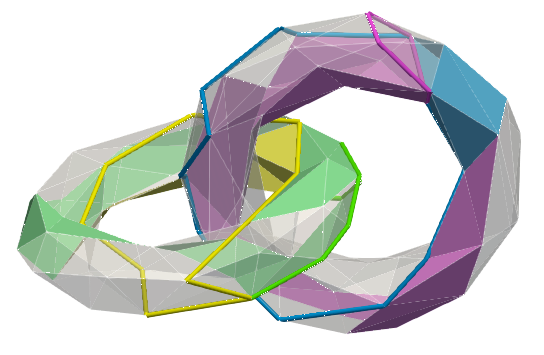
| std::vector< Cell_pair > HDVF::compute_perfect_hdvf | ( | bool | verbose = false | ) |
Computes a perfect HDVF.
As long as valid pairs for A exist, the function selects the first available pair (returned by find_pair_A()) and applies the corresponding A() operation. If the IntegralDomainWithoutDivision of coefficients is a field, this operation always produces a perfect HDVF (ie. the reduced boundary is null and the reduction provides homology and cohomology information).
If the HDVF is initially not trivial (some cells have already been paired), the function completes it into a perfect HDVF.
| verbose | If true, all intermediate reductions are printed out. |
| std::vector< Cell_pair > HDVF::compute_rand_perfect_hdvf | ( | bool | verbose = false | ) |
Computes a random perfect HDVF.
As long as valid pairs for A exist, the function selects a random pair (among pairs returned by find_pairs_A()) and applies the corresponding A() operation. If the IntegralDomainWithoutDivision of coefficients is a field, this operation always produces a perfect HDVF (ie. the reduced boundary is null and the reduction provides homology and cohomology information).
If the HDVF is initially not trivial (some cells have already been paired), the function randomly completes it into a perfect HDVF.
| verbose | If true, all intermediate reductions are printed out. |
|
virtual |
Finds a valid Cell_pair of dimension q / q+1 for A.
The function searches a pair of critical cells \((\gamma_1, \gamma2)\) of dimension q / q+1, valid for A (ie. such that \(\langle \partial_{q+1}(\gamma_2), \gamma_1 \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Lower dimension of the pair. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
|
virtual |
Finds a valid Cell_pair for A containing gamma (a cell of dimension q)
The function searches a cell \(\gamma'\) such that one of the following conditions holds:
| q | Dimension of the cell gamma. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| gamma | Index of a cell to pair. |
|
virtual |
Finds all valid Cell_pair of dimension q / q+1 for A.
The function searches all pairs of critical cells \((\gamma_1, \gamma2)\) of dimension q / q+1, valid for A (ie. such that \(\langle \partial_{q+1}(\gamma_2), \gamma_1 \rangle\) invertible). It returns a vector of such pairs.
| q | Lower dimension of the pair. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
|
virtual |
Finds all valid Cell_pair for A containing gamma (a cell of dimension q)
The function searches all critical cells \(\gamma'\) such that one of the following conditions holds:
| q | Dimension of the cell gamma. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| gamma | Index of a cell to pair. |
| std::vector< std::vector< size_t > > HDVF::flag | ( | PSC_flag | flag | ) | const |
Gets cells with a given PSC_flag in any dimension.
The function returns in each dimension the vector of cells with a given PSC_flag.
|
virtual |
Returns a chain containing the homology generator associated to cell_index (critical cell) of dimension q (in particular for vtk export).
The method exports the chain \(g(\sigma)\) for \(\sigma\) the cell of index cell_index and dimension q.
| bool HDVF::is_perfect_hdvf | ( | ) |
Tests if a HDVF is perfect.
The function returns true if the reduced boundary matrix is null and false otherwise.
| std::vector< size_t > HDVF::psc_flags | ( | PSC_flag | flag, |
| int | q | ||
| ) | const |
Gets cells with a given PSC_flag in dimension q.
The function returns the vector of cells of dimension q with a given PSC_flag.
|
virtual |
Exports primary/secondary/critical labels (in particular for vtk export).
The method exports the labels of every cell in each dimension.
| std::istream & HDVF::read_hdvf_reduction | ( | std::istream & | in | ) |
| std::ostream & HDVF::write_hdvf_reduction | ( | std::ostream & | out | ) |
| std::ostream & HDVF::write_matrices | ( | std::ostream & | out = std::cout | ) | const |
Writes the matrices of the reduction.
Writes the matrices of the reduction (that is \(f\), \(g\), \(h\), \(\partial'\) the reduced boundary).
By default, writes the complex to std::cout.
| std::ostream & HDVF::write_reduction | ( | std::ostream & | out = std::cout | ) | const |
Writes the homology and cohomology reduction information.
Writes the homology and cohomology reduction information (that is \(f^*\), \(g\) \(\partial'\) the reduced boundary over each critical cell).
By default, writes the complex to std::cout.