|
CGAL 6.2 - Homological Discrete Vector Fields
|
|
CGAL 6.2 - Homological Discrete Vector Fields
|
#include <CGAL/HDVF/Hdvf.h>
The class Hdvf implements homology and cohomology computation via homological discrete vector fields (HDVF for short).
It derives from Hdvf_core and shares all its data and methods.
But besides construction operations and methods (using the A() operation), the Hdvf class implements four other HDVF operations: R, M, W and MW together with appropriate "find_pair()" functions. These operations change the HDVF (that is change homology / cohomology generators) and thus provide a convenient tool to move inside the "space of homology/cohomology computations".
R() operation is the "dual" of the A pairing operation (it cancels the pairing and turns back a PRIMARY/SECONDARY pair into a pair of CRITICAL cells)M() operation exchanges a PRIMARY \(\pi\) and a CRITICAL cell \(\gamma\) (under conditions) and modifies the homology generator associated to \(\gamma\) (while preserving is associated cohomology generator)W() operation exchanges a SECONDARY \(\sigma\) and a CRITICAL cell \(\gamma\) (under conditions) and modifies the cohomology generator associated to \(\gamma\) (while preserving is associated homology generator)MW() operation exchanges a PRIMARY \(\pi\) and a SECONDARY cell \(\sigma\) (under conditions). See the introduction to HDVF for more details on this operation.Using appropriate combinations of such operations, one can change a HDVF until corresponding homology or cohomology generators meet a given basis or delineate a hole.
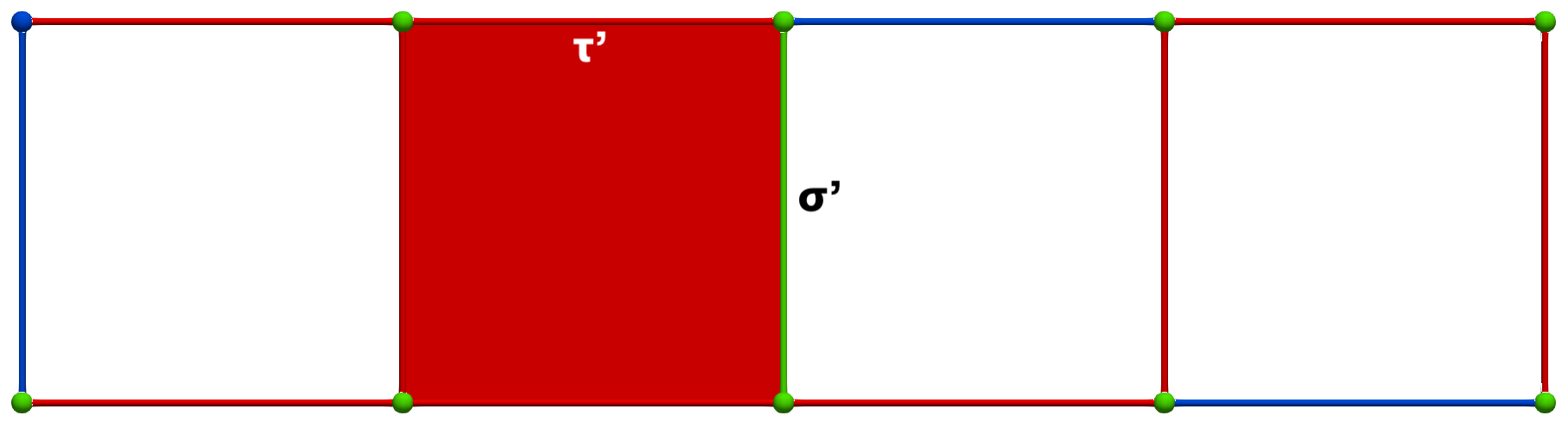
Let us consider the following simple cubical complex and a perfect HDVF (top) together with the three corresponding homology generators (bottom, highlighted in pink):
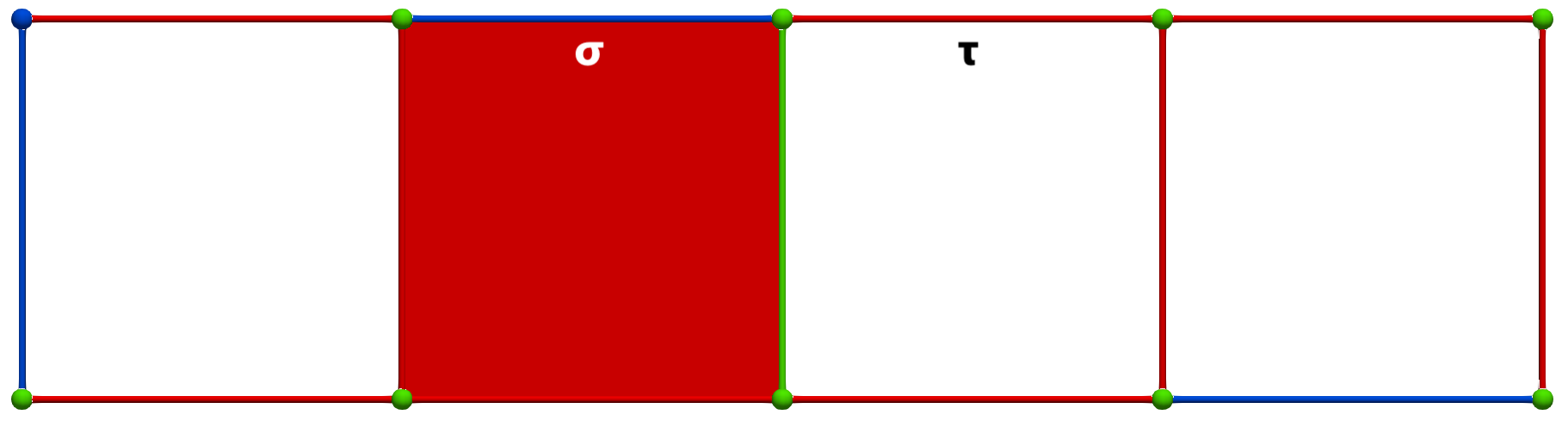
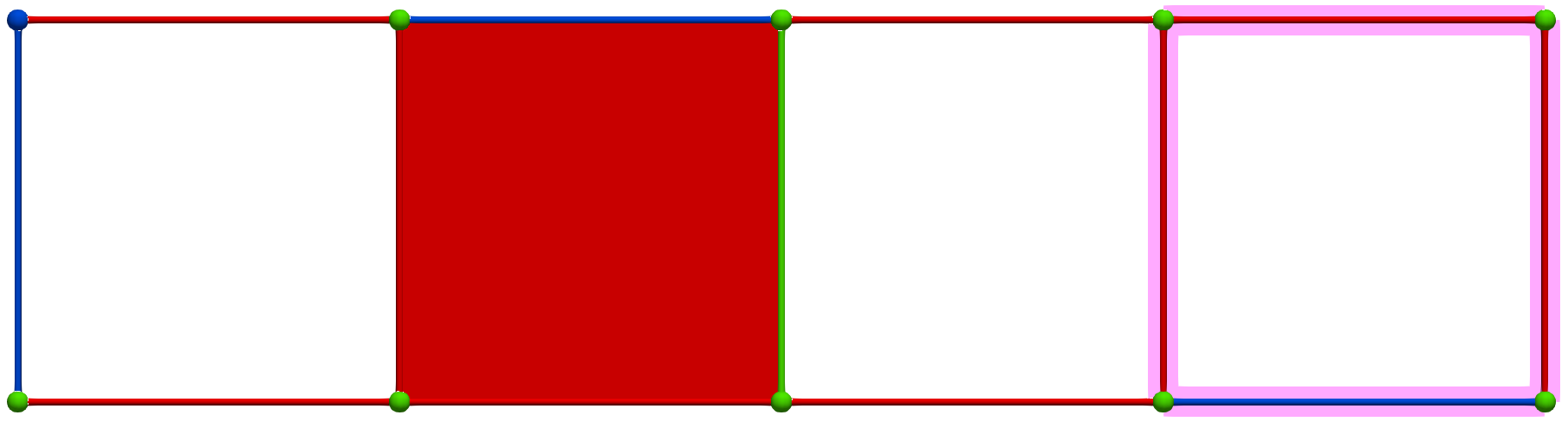
A W operation between cells of Khalimsky coordinates \(\sigma = (3,2)\) (CRITICAL) and \(\tau=(5,2)\) (SECONDARY) produces the following HDVF (homology generators did not change; note that cohomology generators are modified):
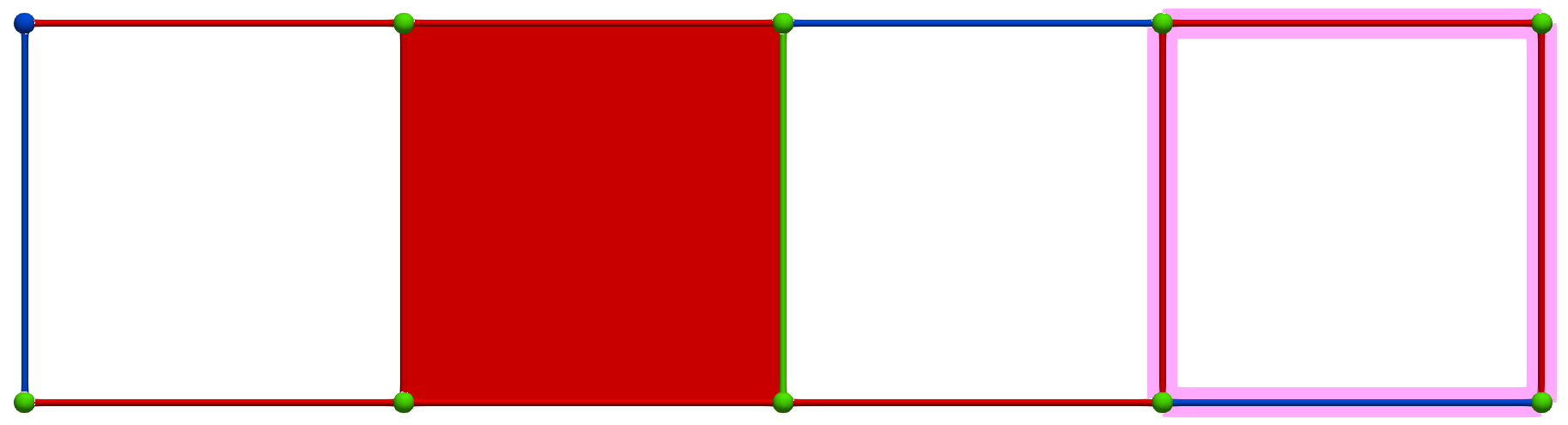
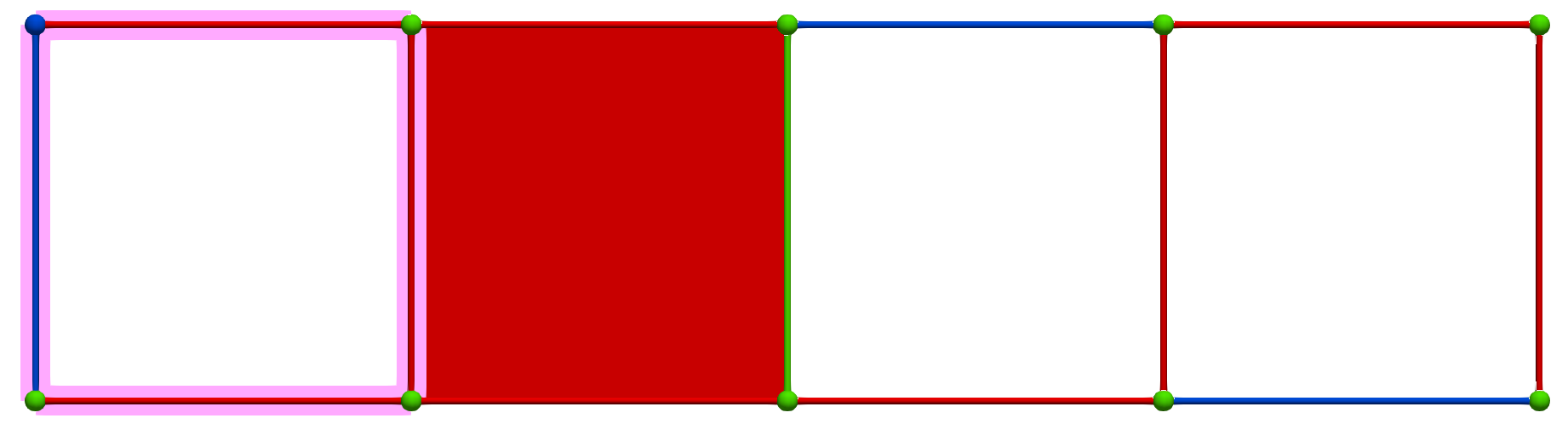
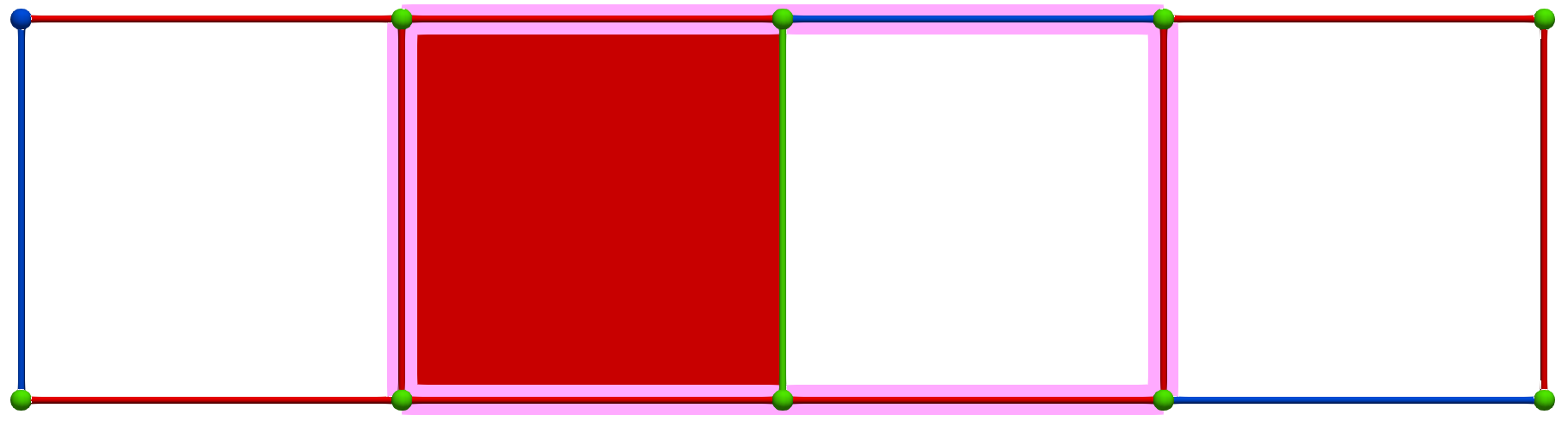
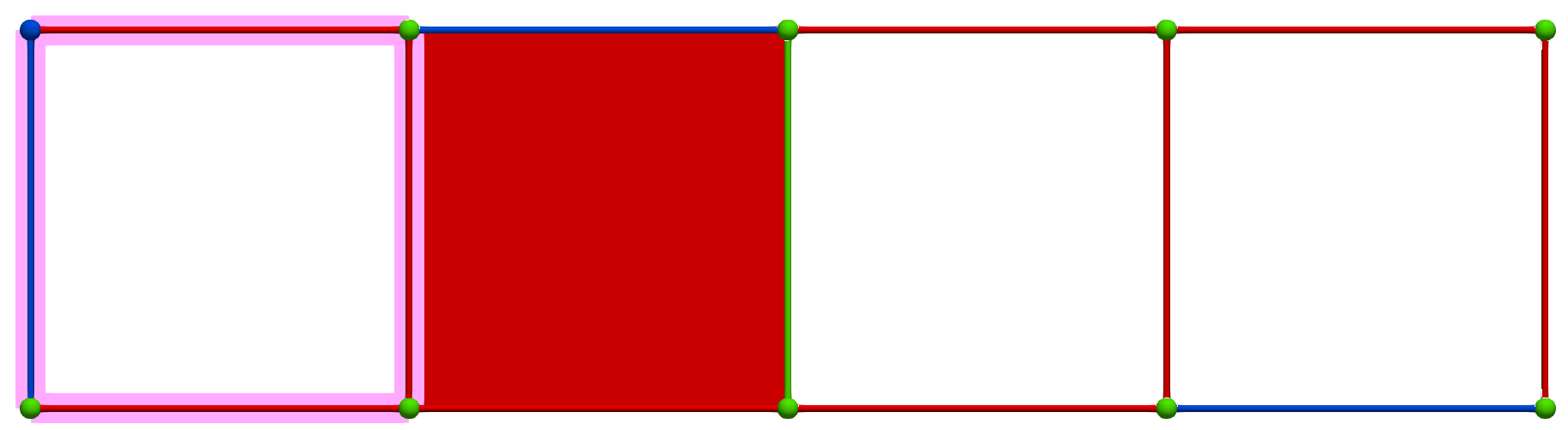
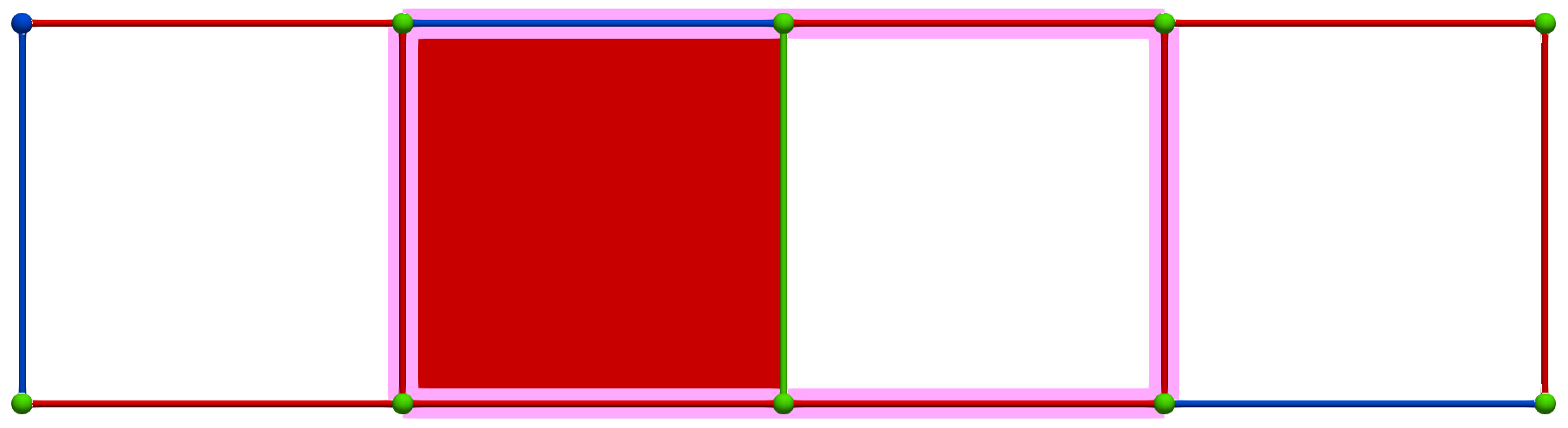
Then, a MW operation between cells of Khalimsky coordinates \(\sigma' = (4,1)\) (PRIMARY) and \(\tau=(3,2)\) (SECONDARY) produces the following HDVF where homology generators become "minimal":
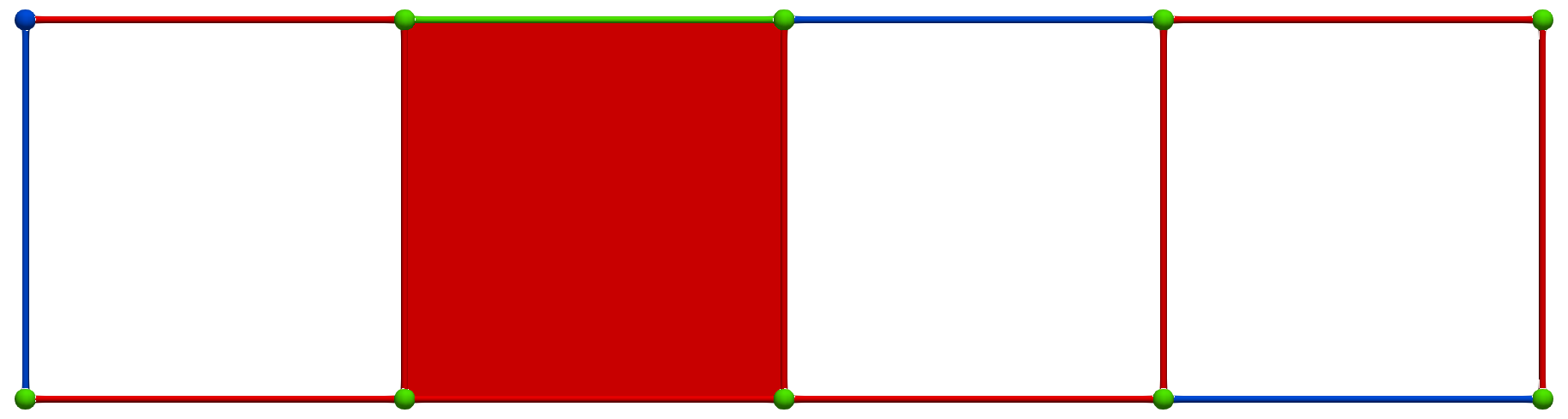
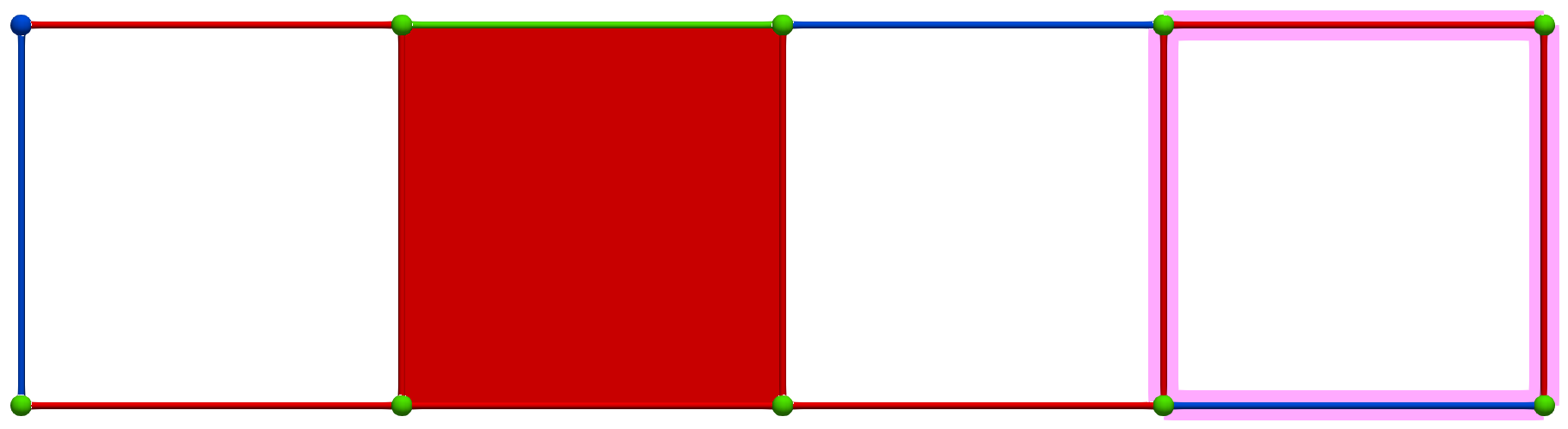
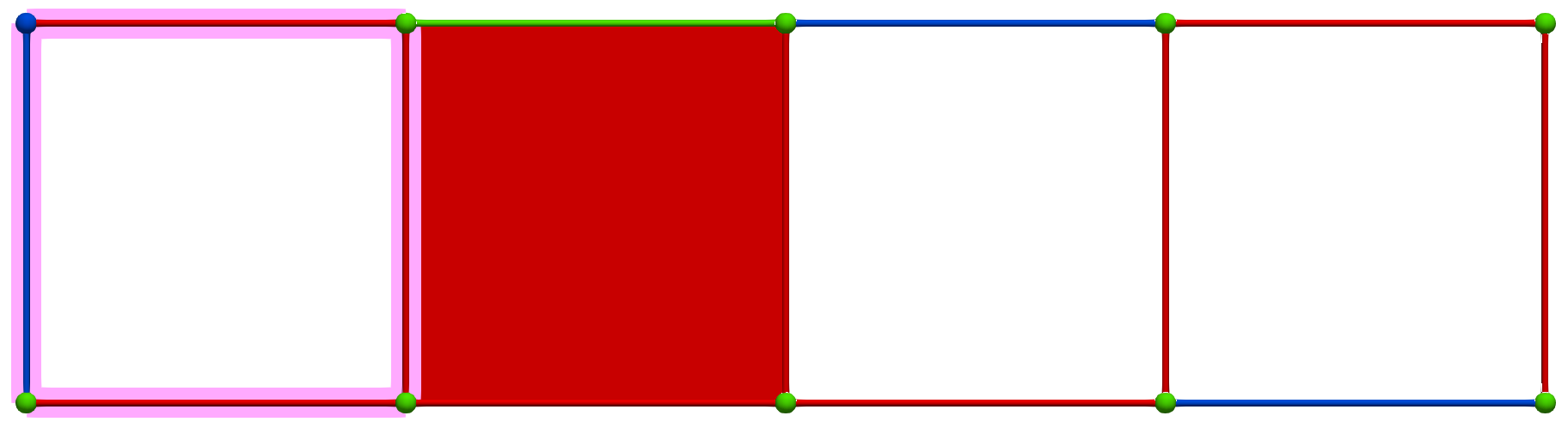
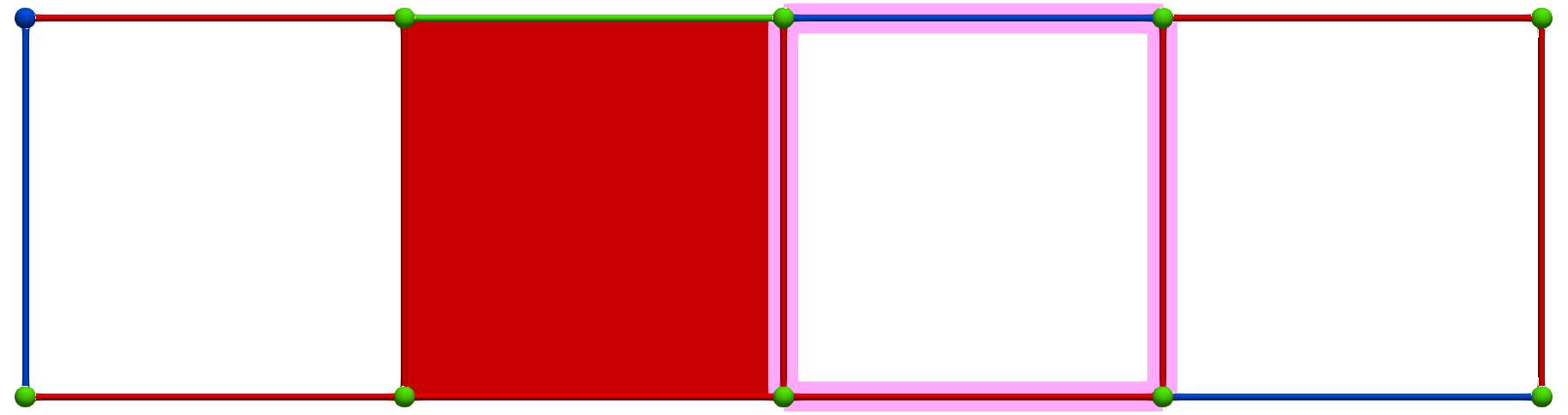
Perfect HDVFs provide various topological results:
psc_flags() with the CRITICALPSC_flag as argument).homology_chain() and cohomology_chain() return the homology and cohomology generator chain associated to a given critical cell. VTK export functions output all the cells of such chains with non zero coefficients. Figures here above illustrate such homology and co-homology generators.get_annotation() or get_coannotation() to get the annotation/co-annotation of a cycle/co-cycle in the homology/cohomology basis (as a chain of critical cells).are_same_cycles() or are_same_cocycles() to check if two cycles (or co-cycles) belong to the same homology / cohomology class.
Figure 94.1 Illustration of cycles annotation and comparison. Left: Generator \(g(\sigma)\) associated to the critical cell \(\sigma\); Middle: A cycle \(\alpha\) ; Right: A second cycle \(\beta\).
For \(\mathbb Z_2\) homology, the annotation of \(\alpha\) is \(\sigma\) ( \(\alpha\) equals \(g(\sigma)\) up to a boundary), while the annotation of \(\beta\) is \(\sigma+\tau+\gamma\) ( \(\beta\) equals \(g(\sigma)+g(\tau)+g(\gamma)\) up to a boundary).
Therefore, are_same_cycles will return true for \(\alpha\) and \(g(\sigma)\), but false for \(\beta\) and \(g(\sigma)\).
HDVF | ChainComplex | a model of the AbstractChainComplex concept, providing the type of abstract chain complex used. |
Public Types | |
| typedef ChainComplex | Chain_complex |
| Chain complex type. | |
| typedef Chain_complex::Coefficient_ring | Coefficient_ring |
| Type of coefficients used to compute homology. | |
| typedef Hdvf_core< Chain_complex, OSM::Sparse_chain, OSM::Sparse_matrix > | Base |
| Type of parent Hdvf_core class. | |
| using | Column_chain = Base::Column_chain |
| using | Row_chain = Base::Row_chain |
| using | Column_matrix = Base::Column_matrix |
| using | Row_matrix = Base::Row_matrix |
 Public Types inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > Public Types inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > | |
| typedef ChainComplex::Coefficient_ring | Coefficient_ring |
| Type of coefficients used to compute homology. | |
| typedef OSM::Sparse_chain< Coefficient_ring, CGAL::OSM::COLUMN > | Column_chain |
| Type of column-major chains. | |
| typedef OSM::Sparse_chain< Coefficient_ring, CGAL::OSM::ROW > | Row_chain |
| Type of row-major chains. | |
| typedef OSM::Sparse_matrix< Coefficient_ring, CGAL::OSM::COLUMN > | Column_matrix |
| Type of column-major sparse matrices. | |
| typedef OSM::Sparse_matrix< Coefficient_ring, CGAL::OSM::ROW > | Row_matrix |
| Type of row-major sparse matrices. | |
Public Member Functions | |
| Hdvf (const Chain_complex &K, int hdvf_opt=OPT_FULL, int dimension_restriction=-1) | |
| Default constructor. | |
| Hdvf (const Hdvf &hdvf) | |
| ~Hdvf () | |
| Cell_pair | find_pair_M (int q, bool &found) const |
| Finds a valid Cell_pair of dimension q for M. | |
| Cell_pair | find_pair_M (int q, bool &found, size_t tau) const |
Finds a valid Cell_pair of dimension q for M cointaining tau. | |
| std::vector< Cell_pair > | find_pairs_M (int q, bool &found) const |
| Finds all valid Cell_pair of dimension q for M. | |
| std::vector< Cell_pair > | find_pairs_M (int q, bool &found, size_t tau) const |
Finds all valid Cell_pair of dimension q for M cointaining tau. | |
| Cell_pair | find_pair_W (int q, bool &found) const |
| Finds a valid Cell_pair of dimension q for W. | |
| Cell_pair | find_pair_W (int q, bool &found, size_t tau) const |
Finds a valid Cell_pair of dimension q for W cointaining tau. | |
| std::vector< Cell_pair > | find_pairs_W (int q, bool &found) const |
| Finds all valid Cell_pair of dimension q for W. | |
| std::vector< Cell_pair > | find_pairs_W (int q, bool &found, size_t tau) const |
Finds all valid Cell_pair of dimension q for W cointaining tau. | |
| Cell_pair | find_pair_MW (int q, bool &found) const |
| Finds a valid Cell_pair of dimension q for MW. | |
| Cell_pair | find_pair_MW (int q, bool &found, size_t tau) const |
Finds a valid Cell_pair of dimension q for MW cointaining tau. | |
| std::vector< Cell_pair > | find_pairs_MW (int q, bool &found) const |
| Finds all valid Cell_pair of dimension q for MW. | |
| std::vector< Cell_pair > | find_pairs_MW (int q, bool &found, size_t tau) const |
Finds all valid Cell_pair of dimension q for W cointaining tau. | |
| void | R (size_t pi, size_t sigma, int q) |
| R operation (cancels a A operation). | |
| void | M (size_t pi, size_t gamma, int q) |
| M operation. | |
| void | W (size_t sigma, size_t gamma, int q) |
| W operation. | |
| void | MW (size_t pi, size_t sigma, int q) |
| MW operation. | |
| Column_chain | get_annotation (Column_chain chain, int dim) const |
| Gets the annotation of a cycle in the homology basis. | |
| Row_chain | get_coannotation (Row_chain chain, int dim) const |
| Gets the co-annotation of a co-cycle in the cohomology basis. | |
| bool | are_same_cycles (Column_chain chain1, Column_chain chain2, int dim) |
| Checks if two cycles belong to the same homology class. | |
| bool | are_same_cocycles (Row_chain chain1, Row_chain chain2, int dim) |
| Checks if two co-cycles belong to the same cohomology class. | |
 Public Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > Public Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > | |
| int | dimension_restriction () const |
| Returns the dimension of Hdvf computation. | |
| void | dimension_restriction (int dimension) |
| Changes the dimension of Hdvf computation. | |
| Hdvf_core (const ChainComplex &K, int hdvf_opt=OPT_FULL, int dimension_restriction=-1) | |
| Constructor from a chain complex. | |
| Hdvf_core (const Hdvf_core &hdvf) | |
| ~Hdvf_core () | |
| virtual Cell_pair | find_pair_A (int q, bool &found) const |
| Finds a valid Cell_pair of dimension q / q+1 for A. | |
| virtual Cell_pair | find_pair_A (int q, bool &found, size_t gamma) const |
Finds a valid Cell_pair for A containing gamma (a cell of dimension q) | |
| virtual std::vector< Cell_pair > | find_pairs_A (int q, bool &found) const |
| Finds all valid Cell_pair of dimension q / q+1 for A. | |
| virtual std::vector< Cell_pair > | find_pairs_A (int q, bool &found, size_t gamma) const |
Finds all valid Cell_pair for A containing gamma (a cell of dimension q) | |
| void | A (size_t gamma1, size_t gamma2, int q) |
| A operation: pairs critical cells. | |
| std::vector< Cell_pair > | compute_perfect_hdvf (bool verbose=false) |
| Computes a perfect HDVF. | |
| std::vector< Cell_pair > | compute_rand_perfect_hdvf (bool verbose=false) |
| Computes a random perfect HDVF. | |
| bool | is_perfect_hdvf (int dimension_restriction=-2) |
| Tests if a HDVF is perfect. | |
| virtual std::vector< std::vector< size_t > > | psc_flags (PSC_flag flag) const |
Gets cells with a given PSC_flag in any dimension. | |
| virtual std::vector< size_t > | psc_flags (PSC_flag flag, int q) const |
Gets cells with a given PSC_flag in dimension q. | |
| PSC_flag | psc_flag (int q, size_t tau) const |
Gets the PSC_flag of the cell tau in dimension q. | |
| int | hdvf_opts () const |
| Gets HDVF computation option. | |
| const Row_matrix & | matrix_f (int q) const |
| Gets the row-major matrix of \(f\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_g (int q) const |
| Gets the column-major matrix of \(g\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_h (int q) const |
| Gets the column-major matrix of \(h\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_dd (int q) const |
| Gets the column-major matrix of \(\partial'\), reduced boundary operator (from the reduction associated to the HDVF). | |
| std::ostream & | write_matrices (std::ostream &out=std::cout, int dimension_restriction=-2) const |
| Writes the matrices of the reduction. | |
| std::ostream & | write_reduction (std::ostream &out=std::cout, int dimension_restriction=-2) const |
| Writes the homology and cohomology reduction information. | |
| virtual std::vector< std::vector< int > > | psc_labels (int dimension_restriction=-2) const |
| Exports primary/secondary/critical integers encoding labels (in particular for vtk export) | |
| virtual Column_chain | homology_chain (size_t cell_index, int q) const |
Gets homology generators associated to cell (critical cell) of dimension q (used by vtk export). | |
| virtual Column_chain | cohomology_chain (size_t cell_index, int dim) const |
Gets cohomology generators associated to cell_index (critical cell) of dimension q (used by vtk export). | |
| std::ostream & | write_hdvf_reduction (std::ostream &out) |
| Writes a HDVF together with the associated reduction (f, g, h, d matrices) | |
| void | write_hdvf_reduction (std::string filename) |
| Writes a HDVF together with the associated reduction to a file (f, g, h, d matrices). | |
| std::istream & | read_hdvf_reduction (std::istream &in_stream) |
| Loads a HDVF together with the associated reduction (f, g, h, d matrices) | |
| void | read_hdvf_reduction (std::string filename) |
| Loads a HDVF together with the associated reduction from a file (f, g, h, d matrices) | |
| bool | compare (const Hdvf_core &other, bool full_compare=false) |
| Compares the HDVF with another HDVF over the same underlying complex. | |
| bool | operator== (const Hdvf_core &other) |
| Comparison operator. | |
Additional Inherited Members | |
 Protected Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > Protected Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > | |
| OSM::Sparse_chain< Coefficient_ring, StorageFormat > | projection (const OSM::Sparse_chain< Coefficient_ring, StorageFormat > &chain, PSC_flag flag, int q) const |
| void | progress_bar (size_t i, size_t n) |
 Protected Attributes inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > Protected Attributes inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sparse_matrix > | |
| std::vector< std::vector< PSC_flag > > | _flag |
| std::vector< size_t > | _nb_P |
| std::vector< size_t > | _nb_S |
| std::vector< size_t > | _nb_C |
| std::vector< Row_matrix > | _F_row |
| std::vector< Column_matrix > | _G_col |
| std::vector< Column_matrix > | _H_col |
| std::vector< Column_matrix > | _DD_col |
| const ChainComplex & | _K |
| int | _hdvf_opt |
| int | _dimension_restriction |
| int | _min_dimension |
| int | _max_dimension |
| CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::Hdvf | ( | const Chain_complex & | K, |
| int | hdvf_opt = OPT_FULL, |
||
| int | dimension_restriction = -1 |
||
| ) |
Default constructor.
Builds an" empty" HDVF associated to K (with all cells critical). By default, the HDVF option is set to OPT_FULL (full reduction computed).
| K | A chain complex (a model of AbstractChainComplex) |
| hdvf_opt | Option for HDVF computation (OPT_BND, OPT_F, OPT_G or OPT_FULL) |
| dimension_restriction | Determines if perfect HDVFs are computed along any dimensions (if dimension_restriction is -1) or a single dimension (specified by dimension_restrictions) |
| bool CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::are_same_cocycles | ( | Row_chain | chain1, |
| Row_chain | chain2, | ||
| int | dim | ||
| ) |
Checks if two co-cycles belong to the same cohomology class.
| chain1 | First co-cycle. |
| chain2 | Second co-cycle. |
| dim | Dimension of both co-cycles. |
| bool CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::are_same_cycles | ( | Column_chain | chain1, |
| Column_chain | chain2, | ||
| int | dim | ||
| ) |
Checks if two cycles belong to the same homology class.
| chain1 | First cycle. |
| chain2 | Second cycle. |
| dim | Dimension of both cycles. |
| Cell_pair CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pair_M | ( | int | q, |
| bool & | found | ||
| ) | const |
Finds a valid Cell_pair of dimension q for M.
The function searches a pair of cells \((\pi, \gamma)\) with \(\pi\) PRIMARY and \(\gamma\) CRITICAL, valid for M (ie. such that \(\langle f(\pi), \gamma \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| Cell_pair CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pair_M | ( | int | q, |
| bool & | found, | ||
| size_t | tau | ||
| ) | const |
Finds a valid Cell_pair of dimension q for M cointaining tau.
The function searches a pair of cells \((\pi, \gamma)\) with \(\pi\) PRIMARY and \(\gamma\) CRITICAL (one of them is tau), valid for M (ie. such that \(\langle f(\pi), \gamma \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| tau | Cell of dimension q to pair. |
| Cell_pair CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pair_MW | ( | int | q, |
| bool & | found | ||
| ) | const |
Finds a valid Cell_pair of dimension q for MW.
The function searches a pair of cells \((\pi, \sigma)\) with \(\pi\) PRIMARY and \(\sigma\) SECONDARY, valid for MW (ie. such that \(\langle h_{q-1}\partial_q(\pi), \sigma \rangle\) invertible and \(\langle \partial_{q+1} h_q(\sigma), \pi \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| Cell_pair CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pair_MW | ( | int | q, |
| bool & | found, | ||
| size_t | tau | ||
| ) | const |
Finds a valid Cell_pair of dimension q for MW cointaining tau.
The function searches a pair of cells \((\pi, \sigma)\) with \(\pi\) PRIMARY and \(\sigma\) SECONDARY (one of them is tau), valid for MW (ie. such that \(\langle h_{q-1}\partial_q(\pi), \sigma \rangle\) invertible and \(\langle \partial_{q+1} h_q(\sigma), \pi \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| tau | Cell of dimension q to pair. |
| Cell_pair CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pair_W | ( | int | q, |
| bool & | found | ||
| ) | const |
Finds a valid Cell_pair of dimension q for W.
The function searches a pair of cells \((\sigma, \gamma)\) with \(\sigma\) SECONDARY and \(\gamma\) CRITICAL, valid for W (ie. such that \(\langle g(\gamma), \sigma \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| Cell_pair CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pair_W | ( | int | q, |
| bool & | found, | ||
| size_t | tau | ||
| ) | const |
Finds a valid Cell_pair of dimension q for W cointaining tau.
The function searches a pair of cells \((\sigma, \gamma)\) with \(\sigma\) SECONDARY and \(\gamma\) CRITICAL (one of them is tau), valid for W (ie. such that \(\langle g(\gamma), \sigma \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| tau | Cell of dimension q to pair. |
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pairs_M | ( | int | q, |
| bool & | found | ||
| ) | const |
Finds all valid Cell_pair of dimension q for M.
The function searches all pairs of cells \((\pi, \gamma)\) with \(\pi\) PRIMARY and \(\gamma\) CRITICAL, valid for M (ie. such that \(\langle f(\pi), \gamma \rangle\) invertible). It returns a vector of such pairs.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pairs_M | ( | int | q, |
| bool & | found, | ||
| size_t | tau | ||
| ) | const |
Finds all valid Cell_pair of dimension q for M cointaining tau.
The function searches all pairs of cells \((\pi, \gamma)\) with \(\pi\) PRIMARY and \(\gamma\) CRITICAL (one of them is tau), valid for M (ie. such that \(\langle f(\pi), \gamma \rangle\) invertible). It returns a vector of such pairs.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| tau | Cell of dimension q to pair. |
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pairs_MW | ( | int | q, |
| bool & | found | ||
| ) | const |
Finds all valid Cell_pair of dimension q for MW.
The function searches all pairs of cells \((\pi, \sigma)\) with \(\pi\) PRIMARY and \(\sigma\) SECONDARY, valid for MW (ie. such that \(\langle h_{q-1}\partial_q(\pi), \sigma \rangle\) invertible and \(\langle \partial_{q+1} h_q(\sigma), \pi \rangle\) invertible). It returns a vector of such pairs.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pairs_MW | ( | int | q, |
| bool & | found, | ||
| size_t | tau | ||
| ) | const |
Finds all valid Cell_pair of dimension q for W cointaining tau.
The function searches all pairs of cells \((\pi, \sigma)\) with \(\pi\) PRIMARY and \(\sigma\) SECONDARY (one of them is tau), valid for MW (ie. such that \(\langle h_{q-1}\partial_q(\pi), \sigma \rangle\) invertible and \(\langle \partial_{q+1} h_q(\sigma), \pi \rangle\) invertible). It returns a vector of such pairs.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| tau | Cell of dimension q to pair. |
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pairs_W | ( | int | q, |
| bool & | found | ||
| ) | const |
Finds all valid Cell_pair of dimension q for W.
The function searches all pairs of cells \((\sigma, \gamma)\) with \(\sigma\) SECONDARY and \(\gamma\) CRITICAL, valid for W (ie. such that \(\langle g(\gamma), \sigma \rangle\) invertible). It returns a vector of such pairs.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::find_pairs_W | ( | int | q, |
| bool & | found, | ||
| size_t | tau | ||
| ) | const |
Finds all valid Cell_pair of dimension q for W cointaining tau.
The function searches all pairs of cells \((\sigma, \gamma)\) with \(\sigma\) SECONDARY and \(\gamma\) CRITICAL (one of them is tau), valid for W (ie. such that \(\langle g(\gamma), \sigma \rangle\) invertible). It returns the first valid pair found by iterators. It returns a vector of such pairs.
| q | Dimension of the pair searched. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| tau | Cell of dimension q to pair. |
| Column_chain CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::get_annotation | ( | Column_chain | chain, |
| int | dim | ||
| ) | const |
Gets the annotation of a cycle in the homology basis.
The method returns the image of a cycle by the morphism \(f\), that is, a linear combination of critical cells (corresponding to the decomposition of the cycle in the homology basis). If the annotation has a single non zero coefficient for a given critical cell \(\sigma\), then the cycle belongs to the class of the homology generator \(g(\sigma)\).
| chain | The cycle to annotate in the homology basis. |
| dim | Dimension of the cycle. |
| Row_chain CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::get_coannotation | ( | Row_chain | chain, |
| int | dim | ||
| ) | const |
Gets the co-annotation of a co-cycle in the cohomology basis.
The method returns the image of a co-cycle by the morphism \(g^*\), that is, a linear combination of critical cells (corresponding to the decomposition of the co-cycle in the cohomology basis). If the annotation has a single non zero coefficient for a given critical cell \(\sigma\), then the co-cycle belongs to the class of the cohomology generator \(f^*(\sigma)\).
| chain | The co-cycle to annotate in the homology basis. |
| dim | Dimension of the co-cycle. |
| void CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::M | ( | size_t | pi, |
| size_t | gamma, | ||
| int | q | ||
| ) |
M operation.
A pair of cells \((\pi, \gamma)\) of dimension q, with \(\pi\) PRIMARY and \(\gamma\) CRITICAL, is valid for M if \(\langle f(\pi), \gamma \rangle\) is invertible. After the M operation, \(\pi\) becomes CRITICAL and \(\gamma\) become PRIMARY. The M method updates the reduction accordingly (in time \(\mathcal O(n^2)\)).
| pi | First cell of the pair (dimension q) |
| gamma | Second cell of the pair (dimension q) |
| q | Dimension of the pair |
| void CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::MW | ( | size_t | pi, |
| size_t | sigma, | ||
| int | q | ||
| ) |
MW operation.
A pair of cells \((\pi, \sigma)\) of dimension q, with \(\pi\) PRIMARY and \(\sigma\) SECONDARY, is valid for MW if \(\langle h_{q-1}\partial_q(\pi), \sigma \rangle\) is invertible and \(\langle \partial_{q+1} h_q(\sigma), \pi \rangle\) is invertible. After the MW operation, \(\pi\) becomes SECONDARY and \(\sigma\) become PRIMARY. The MW method updates the reduction accordingly (in time \(\mathcal O(n^2)\)).
| pi | First cell of the pair (dimension q) |
| sigma | Second cell of the pair (dimension q) |
| q | Dimension of the pair |
| void CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::R | ( | size_t | pi, |
| size_t | sigma, | ||
| int | q | ||
| ) |
R operation (cancels a A operation).
A pair of cells \((\pi, \sigma)\) of respective dimension q and q+1, with \(\pi\) PRIMARY and \(\sigma\) SECONDARY, is valid for R if \(\langle h(\pi), \sigma \rangle\) is invertible. After the R operation, \(\pi\) and \(\sigma\) become CRITICAL. The R method updates the reduction accordingly (in time \(\mathcal O(n^2)\)).
| pi | First cell of the pair (dimension q) |
| sigma | Second cell of the pair (dimension q+1) |
| q | Dimension of the pair |
| void CGAL::Homological_discrete_vector_field::Hdvf< ChainComplex >::W | ( | size_t | sigma, |
| size_t | gamma, | ||
| int | q | ||
| ) |
W operation.
A pair of cells \((\sigma, \gamma)\) of dimension q, with \(\sigma\) SECONDARY and \(\gamma\) CRITICAL, is valid for W if \(\langle g(\gamma), \sigma \rangle\) is invertible. After the W operation, \(\sigma\) becomes CRITICAL and \(\gamma\) become SECONDARY. The W method updates the reduction accordingly (in time \(\mathcal O(n^2)\)).
| sigma | First cell of the pair (dimension q) |
| gamma | Second cell of the pair (dimension q) |
| q | Dimension of the pair |