|
CGAL 6.2 - Homological Discrete Vector Fields
|
|
CGAL 6.2 - Homological Discrete Vector Fields
|
#include <CGAL/HDVF/Hdvf_duality.h>
The class Hdvf_duality is the implementation of homological discrete vector fields (HDVF for short) for Alexander duality computation.
In dimension \(n\), given a complex \(L\) homeomorphic to \(\mathcal S^n\) and a sub-complex \(K\subseteq L\), Alexander duality states that for all \(q\leqslant n\):
\[\tilde H_q(K) \simeq \tilde H^{n-q-1}(L-K)\]
where \(\tilde H_q\) and \(\tilde H^q\) denote reduced homology and cohomology groups.
In [Gonzalez and al. 2025], the authors prove that, even if \(L-K\) is not a sub-complex, it produces a valid chain complex (we call "co-complex" such a complementary of sub-complex). Hence, its homology/cohomology can be computed and for all \(q\leqslant n\):
\[\tilde H_q(K) \simeq \tilde H^{q+1}(L-K)\]
HDVFs provide a fast and convenient mean to compute this isomorphism. In order to work with convenient finite complexes, the complex \(L\) must be homeomorphic to a ball of dimension \(n\) (thus \(\mathcal S^n\) is actually homeomorphic to \(L\) plus an infinite \(n\)-cell closing its boundary).
Perfect HDVFs are first computed over \(K\) and \(L-K\) (providing corresponding relative homology) respectively and Alexander isomorphism gives rise to a pairing between critical cells in \(K\) and \(L-K\), that is a pairing between homology/cohomology generators in \(K\) and \(L-K\).
The class provides HDVF constuction operations: compute_perfect_hdvf() and compute_rand_perfect_hdvf(), which build perfect HDVFs over \(K\) and \(L-K\) respectively. Then, compute_alexander_pairing() computes Alexander isomorphism (and provides a pairing between homology/cohomology generators in \(K\) and \(L-K\)).

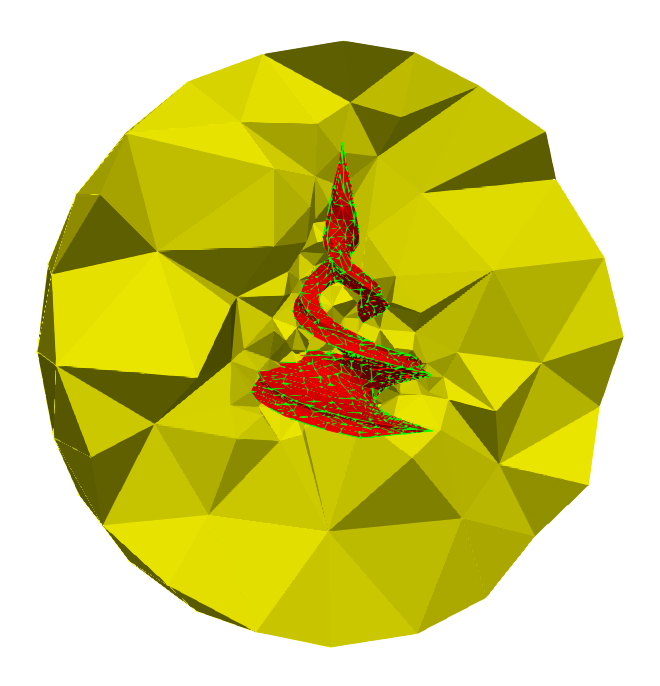
Example of Alexander duality isomorphism. The twirl mesh is a subcomplex K of a larger complex L depicted in yellow, homeomorphic to the ball of dimension 3 (right - sectional view).

Figure 94.1 Example of "homological quartet for the twirl model". 1: Homology generators of the twirl \(H_1(K)\), 2: Cohomology generators of the twirl \(H^1(K)\), 3: Homology generators of the complementary of the twirl \(H_1(L-K)\), 4: Cohomology generators of the complementary of the twirl \(H^1(L-K)\). Alexander isomorphism is represented through colours (paired generators have similar colours).
Hence, each hole in \(K\) gives rise to four generators (called its "homological quarted": its homology and cohomology generators in \(K_q\) and the homology and cohomology generators paired with them in \((L-K)_{q+1}\)).
In order to compute relative homology, a sub chain complex mask is used to partially screen the complex L and thus restrict HDVF computation. This mask is called "current mask" (and can be set over K or L-K).
HDVF | ChainComplex | a model of the AbstractChainComplex concept, providing the type of abstract chain complex used. |
[Gonzalez and al. 2025] Gonzalez-Lorenzo, A., Bac, A. & Gazull, YS. A constructive approach of Alexander duality. J Appl. and Comput. Topology 9, 2 (2025).
Public Types | |
| typedef ChainComplex | Chain_complex |
| Chain complex type. | |
| typedef Chain_complex::Coefficient_ring | Coefficient_ring |
| Type of coefficients used to compute homology. | |
| typedef Hdvf_core< ChainComplex, CGAL::OSM::Sparse_chain, CGAL::OSM::Sub_sparse_matrix > | Base |
| Type of parent Hdvf_core class. | |
| using | Column_chain = Base::Column_chain |
| using | Row_chain = Base::Row_chain |
 Public Types inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Public Types inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| typedef ChainComplex::Coefficient_ring | Coefficient_ring |
| Type of coefficients used to compute homology. | |
| typedef OSM::Sparse_chain< Coefficient_ring, CGAL::OSM::COLUMN > | Column_chain |
| Type of column-major chains. | |
| typedef OSM::Sparse_chain< Coefficient_ring, CGAL::OSM::ROW > | Row_chain |
| Type of row-major chains. | |
| typedef OSM::Sub_sparse_matrix< Coefficient_ring, CGAL::OSM::COLUMN > | Column_matrix |
| Type of column-major sparse matrices. | |
| typedef OSM::Sub_sparse_matrix< Coefficient_ring, CGAL::OSM::ROW > | Row_matrix |
| Type of row-major sparse matrices. | |
Public Member Functions | |
| Hdvf_duality (const Chain_complex &L, Sub_chain_complex_mask< Chain_complex > &K, int hdvf_opt=OPT_FULL) | |
Hdvf_duality constructor ( from a complex L and a sub-complex K) | |
| Cell_pair | find_pair_A (int q, bool &found) const |
| Finds a valid cell pair of dimension q / q+1 for A in the current sub chain complex. | |
| Cell_pair | find_pair_A (int q, bool &found, size_t gamma) const |
Finds a valid cell pair for A containing gamma in the current sub chain complex (a cell of dimension q) | |
| std::vector< Cell_pair > | find_pairs_A (int q, bool &found) const |
| Finds all valid cell pairs of dimension q / q+1 in the current sub chain complex for A. | |
| std::vector< Cell_pair > | find_pairs_A (int q, bool &found, size_t gamma) const |
Finds all valid cell pairs for Acontaining gamma in the current sub chain complex (a cell of dimension q) | |
| void | set_mask_K () |
Sets the current sub chain complex masks over K. | |
| void | set_mask_L_K () |
Sets the current sub chain complex masks over L-K. | |
| Sub_chain_complex_mask< ChainComplex > | get_current_mask () |
| Returns the value of the current sub chain complex mask. | |
| std::vector< Cell_pair > | compute_perfect_hdvf (bool verbose=false) |
| Computes a perfect HDVF over the current sub chain complex. | |
| std::vector< Cell_pair > | compute_rand_perfect_hdvf (bool verbose=false) |
| Computes a random perfect HDVF over the current sub chain complex. | |
| std::vector< Cell_pair > | compute_pairing_hdvf () |
| Computes a "pairing" HDVF between K and L-K. | |
| std::vector< Cell_pair > | compute_rand_pairing_hdvf () |
| Computes a random "pairing" HDVF between K and L-K. | |
| std::vector< std::vector< size_t > > | psc_flags (PSC_flag flag) const |
Gets cells with a given PSC_flag in any dimension in the current sub chain complex. | |
| std::vector< size_t > | psc_flags (PSC_flag flag, int q) const |
Gets cells with a given PSC_flag in dimension q in the current sub chain complex. | |
| std::ostream & | insert_reduction (std::ostream &out=std::cout) |
Prints the homology and cohomology reduction information for K and L-K. | |
| std::ostream & | print_reduction_sub (std::ostream &out=std::cout) |
| Prints the homology and cohomology reduction information for the current such chain complex. | |
| std::ostream & | print_bnd_pairing (std::ostream &out=std::cout) |
Prints the reduced boundary over critical cells of K and L-K. | |
| std::vector< std::vector< int > > | psc_labels () const |
| Exports primary/secondary/critical labels of the current sub chain complex for vtk export. | |
| Column_chain | homology_chain (size_t cell_index, int dim) const |
Exports homology generators of the current sub chain complex associated to cell_index (critical cell) of dimension q (used by vtk export). | |
| Column_chain | cohomology_chain (size_t cell_index, int dim) const |
Exports cohomology generators of the current sub chain complex associated to cell_index (critical cell) of dimension q (used by vtk export). | |
 Public Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Public Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| int | dimension_restriction () const |
| Returns the dimension of Hdvf computation. | |
| void | dimension_restriction (int dimension) |
| Changes the dimension of Hdvf computation. | |
| Hdvf_core (const ChainComplex &K, int hdvf_opt=OPT_FULL, int dimension_restriction=-1) | |
| Constructor from a chain complex. | |
| Hdvf_core (const Hdvf_core &hdvf) | |
| ~Hdvf_core () | |
| virtual Cell_pair | find_pair_A (int q, bool &found) const |
| Finds a valid Cell_pair of dimension q / q+1 for A. | |
| virtual Cell_pair | find_pair_A (int q, bool &found, size_t gamma) const |
Finds a valid Cell_pair for A containing gamma (a cell of dimension q) | |
| virtual std::vector< Cell_pair > | find_pairs_A (int q, bool &found) const |
| Finds all valid Cell_pair of dimension q / q+1 for A. | |
| virtual std::vector< Cell_pair > | find_pairs_A (int q, bool &found, size_t gamma) const |
Finds all valid Cell_pair for A containing gamma (a cell of dimension q) | |
| void | A (size_t gamma1, size_t gamma2, int q) |
| A operation: pairs critical cells. | |
| std::vector< Cell_pair > | compute_perfect_hdvf (bool verbose=false) |
| Computes a perfect HDVF. | |
| std::vector< Cell_pair > | compute_rand_perfect_hdvf (bool verbose=false) |
| Computes a random perfect HDVF. | |
| bool | is_perfect_hdvf (int dimension_restriction=-2) |
| Tests if a HDVF is perfect. | |
| virtual std::vector< std::vector< size_t > > | psc_flags (PSC_flag flag) const |
Gets cells with a given PSC_flag in any dimension. | |
| virtual std::vector< size_t > | psc_flags (PSC_flag flag, int q) const |
Gets cells with a given PSC_flag in dimension q. | |
| PSC_flag | psc_flag (int q, size_t tau) const |
Gets the PSC_flag of the cell tau in dimension q. | |
| int | hdvf_opts () const |
| Gets HDVF computation option. | |
| const Row_matrix & | matrix_f (int q) const |
| Gets the row-major matrix of \(f\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_g (int q) const |
| Gets the column-major matrix of \(g\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_h (int q) const |
| Gets the column-major matrix of \(h\) (from the reduction associated to the HDVF). | |
| const Column_matrix & | matrix_dd (int q) const |
| Gets the column-major matrix of \(\partial'\), reduced boundary operator (from the reduction associated to the HDVF). | |
| std::ostream & | write_matrices (std::ostream &out=std::cout, int dimension_restriction=-2) const |
| Writes the matrices of the reduction. | |
| std::ostream & | write_reduction (std::ostream &out=std::cout, int dimension_restriction=-2) const |
| Writes the homology and cohomology reduction information. | |
| virtual std::vector< std::vector< int > > | psc_labels (int dimension_restriction=-2) const |
| Exports primary/secondary/critical integers encoding labels (in particular for vtk export) | |
| virtual Column_chain | homology_chain (size_t cell_index, int q) const |
Gets homology generators associated to cell (critical cell) of dimension q (used by vtk export). | |
| virtual Column_chain | cohomology_chain (size_t cell_index, int dim) const |
Gets cohomology generators associated to cell_index (critical cell) of dimension q (used by vtk export). | |
| std::ostream & | write_hdvf_reduction (std::ostream &out) |
| Writes a HDVF together with the associated reduction (f, g, h, d matrices) | |
| void | write_hdvf_reduction (std::string filename) |
| Writes a HDVF together with the associated reduction to a file (f, g, h, d matrices). | |
| std::istream & | read_hdvf_reduction (std::istream &in_stream) |
| Loads a HDVF together with the associated reduction (f, g, h, d matrices) | |
| void | read_hdvf_reduction (std::string filename) |
| Loads a HDVF together with the associated reduction from a file (f, g, h, d matrices) | |
| bool | compare (const Hdvf_core &other, bool full_compare=false) |
| Compares the HDVF with another HDVF over the same underlying complex. | |
| bool | operator== (const Hdvf_core &other) |
| Comparison operator. | |
Additional Inherited Members | |
 Protected Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Protected Member Functions inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| OSM::Sparse_chain< Coefficient_ring, StorageFormat > | projection (const OSM::Sparse_chain< Coefficient_ring, StorageFormat > &chain, PSC_flag flag, int q) const |
| void | progress_bar (size_t i, size_t n) |
 Protected Attributes inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > Protected Attributes inherited from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix > | |
| std::vector< std::vector< PSC_flag > > | _flag |
| std::vector< size_t > | _nb_P |
| std::vector< size_t > | _nb_S |
| std::vector< size_t > | _nb_C |
| std::vector< Row_matrix > | _F_row |
| std::vector< Column_matrix > | _G_col |
| std::vector< Column_matrix > | _H_col |
| std::vector< Column_matrix > | _DD_col |
| const ChainComplex & | _K |
| int | _hdvf_opt |
| int | _dimension_restriction |
| int | _min_dimension |
| int | _max_dimension |
| typedef ChainComplex CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::Chain_complex |
Chain complex type.
| CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::Hdvf_duality | ( | const Chain_complex & | L, |
| Sub_chain_complex_mask< Chain_complex > & | K, | ||
| int | hdvf_opt = OPT_FULL |
||
| ) |
Hdvf_duality constructor ( from a complex L and a sub-complex K)
L is a complex of a given dimension \(n\) homeomorphic to \(\mathcal B^n\) and K is a sub-complex of L described by a bitboard (cells of K have a bit set to 1, cells of K have a bit set to 0).
Initially, the sub chain complex mask is set to K.
| L | A complex of a given dimension \(n\) homeomorphic to \(\mathcal B^n\). |
| K | A sub complex of L encoded through a bitboard. |
| hdvf_opt | Option for HDVF computation (OPT_BND, OPT_F, OPT_G or OPT_FULL). |
|
virtual |
Exports cohomology generators of the current sub chain complex associated to cell_index (critical cell) of dimension q (used by vtk export).
The method exports the chain \(f^\star(\sigma)\) for \(\sigma\) the cell of index cell_index and dimension q.
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::compute_pairing_hdvf |
Computes a "pairing" HDVF between K and L-K.
compute_perfect_hdvf() first (to build perfect HDVFs over K and L-K respectively).The function computes a perfect HDVF over remaining critical cells. Each pair of cells inserted with the A() operation maps corresponding homology/cohomology generators in the Alexander isomorphism.
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::compute_perfect_hdvf | ( | bool | verbose = false | ) |
Computes a perfect HDVF over the current sub chain complex.
As long as valid pairs for A exist in the current sub chain complex, the function selects the first available pair (returned by find_pair_A()) and applies the corresponding A() operation. If the coefficient ring of coefficients is a field, this operation always produces a perfect HDVF (ie. the reduced boundary is null and the reduction provides homology and cohomology information). Otherwise the operation produces a maximal HDVF with a residual boundary matrix over critical cells.
If the HDVF is initially not trivial (some cells have already been paired), the function completes it into a perfect HDVF.
| verbose | If this parameter is true, all intermediate reductions are printed out. |
Cell_pair paired with A. | std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::compute_rand_pairing_hdvf |
Computes a random "pairing" HDVF between K and L-K.
compute_perfect_hdvf() first (to build perfect HDVFs over K and L-K respectively).The function computes a random perfect HDVF over remaining critical cells. Each pair of cells inserted with the A() operation maps corresponding homology/cohomology generators in the Alexander isomorphism.
| std::vector< Cell_pair > CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::compute_rand_perfect_hdvf | ( | bool | verbose = false | ) |
Computes a random perfect HDVF over the current sub chain complex.
As long as valid pairs for A exist in the current sub chain complex, the function selects a random pair (among pairs returned by find_pairs_A()) and applies the corresponding A() operation. If the coefficient ring is a field, this operation always produces a perfect HDVF (that is the reduced boundary is null and the reduction provides homology and cohomology information).
If the HDVF is initially not trivial (some cells have already been paired), the function randomly completes it into a perfect HDVF.
compute_perfect_hdvf() (finding out all possible valid pairs requires additional time).| verbose | If this parameter is true, all intermediate reductions are printed out. |
|
virtual |
Finds a valid cell pair of dimension q / q+1 for A in the current sub chain complex.
The function searches a pair of critical cells, in the current sub chain complex, \((\gamma_1, \gamma2)\) of dimension q / q+1, valid for A (ie. such that \(\langle \partial_{q+1}(\gamma_2), \gamma_1 \rangle\) invertible). It returns the first valid pair found by iterators.
| q | Lower dimension of the pair. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Finds a valid cell pair for A containing gamma in the current sub chain complex (a cell of dimension q)
The function searches a cell \(\gamma'\) in the current sub chain complex such that one of the following conditions holds:
| q | Dimension of the cell gamma. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| gamma | Index of a cell to pair. |
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Finds all valid cell pairs of dimension q / q+1 in the current sub chain complex for A.
The function searches all pairs of critical cells \((\gamma_1, \gamma2)\) in the current sub chain complex of dimension q / q+1, valid for A (ie. such that \(\langle \partial_{q+1}(\gamma_2), \gamma_1 \rangle\) invertible). It returns a vector of such pairs.
| q | Lower dimension of the pairs. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Finds all valid cell pairs for Acontaining gamma in the current sub chain complex (a cell of dimension q)
The function searches all CRITICAL cells \(\gamma'\) in the current sub chain complex such that one of the following conditions holds:
| q | Dimension of the cell gamma. |
| found | Reference to a Boolean variable. The method sets found to true if a valid pair is found, false otherwise. |
| gamma | Index of a cell to pair. |
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Exports homology generators of the current sub chain complex associated to cell_index (critical cell) of dimension q (used by vtk export).
The method exports the chain \(g(\sigma)\) for \(\sigma\) the cell of index cell_index and dimension q.
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
| std::ostream & CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::insert_reduction | ( | std::ostream & | out = std::cout | ) |
Prints the homology and cohomology reduction information for K and L-K.
Prints \(f^*\), \(g\) \(\partial'\) the reduced boundary over each critical cell.
By default, outputs the complex to std::cout.
| std::ostream & CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::print_bnd_pairing | ( | std::ostream & | out = std::cout | ) |
Prints the reduced boundary over critical cells of K and L-K.
The method prints out the reduced boundary matrix in each dimension, restricted to critical cells of K and L-K (ie. the matrix used to compute Alexander pairing).
compute_perfect_hdvf().By default, outputs the complex to std::cout.
| std::ostream & CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::print_reduction_sub | ( | std::ostream & | out = std::cout | ) |
Prints the homology and cohomology reduction information for the current such chain complex.
Prints \(f^*\), \(g\) \(\partial'\) the reduced boundary over each critical cell.
By default, outputs the complex to std::cout.
|
virtual |
Gets cells with a given PSC_flag in any dimension in the current sub chain complex.
The function returns a vector containing, for each dimension, the vector of cells with a given PSC_flag.
| flag | PSC_flag to select. |
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
|
virtual |
Gets cells with a given PSC_flag in dimension q in the current sub chain complex.
The function returns the vector of cells of dimension q with a given PSC_flag.
| flag | PSC_flag to select. |
| q | Dimension visited. |
Reimplemented from CGAL::Homological_discrete_vector_field::Hdvf_core< ChainComplex, OSM::Sparse_chain, OSM::Sub_sparse_matrix >.
| std::vector< std::vector< int > > CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::psc_labels | ( | ) | const |
Exports primary/secondary/critical labels of the current sub chain complex for vtk export.
The method exports the labels of every cells in each dimension.
| void CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::set_mask_K | ( | ) |
Sets the current sub chain complex masks over K.
Further HDVF computations will be restricted to K (ie. computation of reduced homology).
| void CGAL::Homological_discrete_vector_field::Hdvf_duality< ChainComplex >::set_mask_L_K | ( | ) |
Sets the current sub chain complex masks over L-K.
Further HDVF computations will be restricted to L-K (ie. computation of reduced homology).